

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
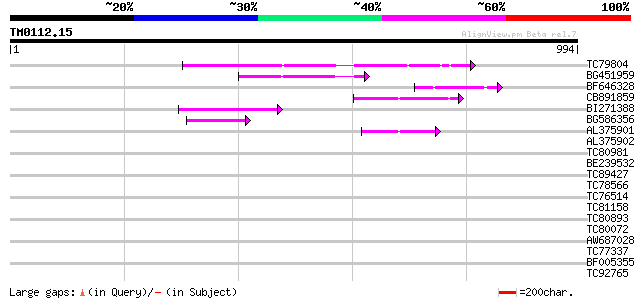
Query= TM0112.15
(994 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like... 207 1e-53
BG451959 homologue to GP|6175165|gb| Mutator-like transposase {A... 106 5e-23
BF646328 similar to PIR|F84533|F845 Mutator-like transposase [im... 80 3e-15
CB891859 similar to GP|6175165|gb| Mutator-like transposase {Ara... 73 6e-13
BI271388 similar to GP|10177186|db mutator-like transposase-like... 71 2e-12
BG586356 similar to GP|9759134|dbj mutator-like transposase-like... 57 5e-08
AL375901 similar to GP|9759134|dbj mutator-like transposase-like... 50 3e-06
AL375902 similar to GP|9759134|dbj mutator-like transposase-like... 41 0.002
TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|... 39 0.013
BE239532 34 0.24
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 33 0.71
TC78566 similar to GP|20466388|gb|AAM20511.1 putative chloroplas... 33 0.71
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 33 0.71
TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidops... 33 0.71
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 33 0.71
TC80072 similar to GP|14423520|gb|AAK62442.1 Unknown protein {Ar... 32 0.92
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 32 0.92
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 32 1.2
BF005355 similar to GP|9758230|dbj| emb|CAB61996.1~gene_id:MZF18... 31 2.7
TC92765 similar to EGAD|119550|127787 hypothetical protein F49E1... 31 2.7
>TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like
transposase {Arabidopsis thaliana}, partial (57%)
Length = 2129
Score = 207 bits (528), Expect = 1e-53
Identities = 146/518 (28%), Positives = 241/518 (46%), Gaps = 5/518 (0%)
Frame = +1
Query: 304 EEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVA-GCPFYLRLSKT 362
E+ F VG F F+ A+K+ A+ Q +L K D R C + GCP+ +R K
Sbjct: 163 EDHNFVVGQEFPDVKAFRNAIKEAAIAQHFELRIIKSDLIRYFAKCASEGCPWRIRAVKL 342
Query: 363 DSRSYWLLVSLEADHTCCRSASN--RQAKTKYLARKFMPIIRHTPNITVNALIEEARLRW 420
+ S + + SLE HTC R+A N QA ++ +R N ++ + ++
Sbjct: 343 PNASTFTIRSLEGTHTCGRNALNGHHQASVDWIVSFIEERLRDNINYKPKDILHDIHKQY 522
Query: 421 GILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVF 480
GI + +A+RAK + + I G + + + L + E+ ++NPGS + +T GA F
Sbjct: 523 GITIPYKQAWRAKERGLAAIYGSSEEGFYLLPSFCEEIKKTNPGSVAKVFTT-GADSR-F 696
Query: 481 ERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAE 540
+R+++ F A+ F C P++ L G LK Y L+A D + +FP+AFAVV+ E
Sbjct: 697 QRLFISFYASIHGFVNGCLPIVALGGIQLKSKYLSTFLSATSFDADGGLFPLAFAVVDVE 876
Query: 541 TKASWSWFVDLLVNDLDMVER--KRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLY 598
SW+WF+ L N L++ + F+SD QK
Sbjct: 877 NDESWTWFLSELHNALEVNTECMPQIIFLSDGQK-------------------------- 978
Query: 599 SCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAM 658
G+V + H C++HL N K++ + + LW AA A+T+ + M
Sbjct: 979 -----GIVDAIRRKFPRSSHAFCMRHLSENIGKEFKNSRLIHLLWSAAYATTINAFREKM 1143
Query: 659 EVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLL 718
++E++ A + + P W + L N E FN+ ILE ++ PII ++
Sbjct: 1144AEIEEVSPNASMWLQHFHPSQWALVYFEGTRYGHLSSN--IEEFNKWILEAQELPIIQVI 1317
Query: 719 ERLKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSR 778
ER++ L ++ W + L P ++++ ++ N + DE FEV
Sbjct: 1318ERIQSKLKTEFDDRRLKSSSWCSVLTPSSERRMVEAI-NRASTYQVLKSDEVE--FEVIS 1488
Query: 779 DNEKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRR 816
+ VN+ S SC+CR W L GIPC+HAVA++ R+
Sbjct: 1489ADRSDIVNIGSHSCSCRDWQLYGIPCSHAVAALISSRK 1602
>BG451959 homologue to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (28%)
Length = 658
Score = 106 bits (264), Expect = 5e-23
Identities = 68/233 (29%), Positives = 103/233 (44%), Gaps = 2/233 (0%)
Frame = +2
Query: 401 IRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLR 460
+ PN ++EE GI L +A+R K + ++G + Y L +Y A + R
Sbjct: 56 LNENPNCKPKEILEEIHRVHGITLSYKQAWRGKEHIMAAMRGSFEEGYRLLPQYCAHVKR 235
Query: 461 SNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAA 520
+NPGS + F+R+++ F A+ CRPL+GLD +LK Y G LL A
Sbjct: 236 TNPGSIASVYGNPSDN--CFQRLFISFQASIYGLLNACRPLLGLDRIYLKSKYLGTLLLA 409
Query: 521 VGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVN--DLDMVERKRWAFISDQQKVMLN*S 578
G DG+ +FP+AF VV+ E +W WF+ L N +++ R +SD+Q
Sbjct: 410 TGFDGDGALFPLAFGVVDEENDDNWMWFLSKLHNLLEINTENMPRLTILSDRQ------- 568
Query: 579 CIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRK 631
QG+V + H C++HL NFRK
Sbjct: 569 ------------------------QGIVDGVEANFPTAFHGFCMRHLSDNFRK 655
>BF646328 similar to PIR|F84533|F845 Mutator-like transposase [imported] -
Arabidopsis thaliana, partial (3%)
Length = 636
Score = 80.5 bits (197), Expect = 3e-15
Identities = 52/158 (32%), Positives = 77/158 (47%), Gaps = 3/158 (1%)
Frame = -1
Query: 710 RDKPIITLLERLKFYLNNRIVKQKQLM--LRWRTALCPMIQQKLEDSKRNSDK-WFANWM 766
R+KP+I LLE + N + K ++W + K+ S + W A W
Sbjct: 630 REKPVIXLLEGYQALYNCEDCQAKADA*KVQW*DM---SKDSSFD*EKQKSCRGWKATWH 460
Query: 767 GDEYMSLFEVSRDNEKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
GD M+ F VS + KY VNL R+CACR+W+L+GIPCAH + +W E++V S Y
Sbjct: 459 GDMEMNNFNVSNETNKYIVNLAQRTCACRKWNLTGIPCAHVIPCIWHNGLADENFVSSYY 280
Query: 827 RHLLIHTPTSSYLIMDLSYGLKFQLLH*THLT*GELRV 864
R + S+ I+ Y + L+ +L G V
Sbjct: 279 R*CSV----STIFILSKMYFYAYMLIWYQYLVAGNHNV 178
>CB891859 similar to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (27%)
Length = 625
Score = 72.8 bits (177), Expect = 6e-13
Identities = 46/193 (23%), Positives = 92/193 (46%)
Frame = +1
Query: 603 QGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMK 662
QG+V + H C++HL +FRK++ + LW AA T+ E+ + ++
Sbjct: 58 QGIVDGVEANFPTAFHGFCMRHLSDSFRKEFNNTMLVNLLWEAANCLTIIEFEGKVMEIE 237
Query: 663 ELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLK 722
E+++ A + +PP++W +AY + N+ EA N ILE PII ++E ++
Sbjct: 238 EISQDAAYWIRRVPPRLWA-TAYFEGHRFGHLTANIVEALNSWILEASGLPIIQMMECIR 414
Query: 723 FYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEK 782
L ++++ ++W + L P ++ + ++ + + + + FEV
Sbjct: 415 RQLMTWFNERRETSMQWTSILVPSAERSVAEALERARTYQVLRANE---AEFEVISHEGT 585
Query: 783 YSVNLESRSCACR 795
V++ +R C CR
Sbjct: 586 NIVDIRNRCCLCR 624
>BI271388 similar to GP|10177186|db mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (26%)
Length = 711
Score = 71.2 bits (173), Expect = 2e-12
Identities = 49/186 (26%), Positives = 85/186 (45%), Gaps = 3/186 (1%)
Frame = +2
Query: 296 RFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVA-GCP 354
R +A Q E V F+S F++A+ Y++ +KK+D R V C + GCP
Sbjct: 95 RQRAAQQWENTITGVDQRFNSFSEFREALHKYSIAHGFAYRYKKNDSHRVTVKCKSQGCP 274
Query: 355 FYLRLSKTDSRSYWLLVSLEADHTCCRSA--SNRQAKTKYLARKFMPIIRHTPNITVNAL 412
+ + SK + + + DHTC SA + +A ++ ++ +PN +
Sbjct: 275 WRIYASKLSTTQLICIKKMTRDHTCEGSAVKAGYRATRGWVGNIIKEKLKASPNYRPKDI 454
Query: 413 IEEARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKST 472
++ + +GI L +A+RAK A E +QG + Y L + ++ +NPG I
Sbjct: 455 ADDIKREYGIQLNYSQAWRAKEIAREQLQGSYKEAYTQLPFFCEKIKETNPGEFCNIHDK 634
Query: 473 KGAKGP 478
+ K P
Sbjct: 635 RXLKFP 652
>BG586356 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 696
Score = 56.6 bits (135), Expect = 5e-08
Identities = 31/116 (26%), Positives = 59/116 (50%), Gaps = 3/116 (2%)
Frame = +3
Query: 310 VGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVA-GCPFYLRLSKTDSRSYW 368
+G F + ++ +KD A+ DL K D+ R + C GCP+ + ++K +
Sbjct: 333 IGQEFPDVETCRRTLKDIAIAMHFDLRIVKSDRSRFIAKCSKEGCPWRVHVAKCPGVPTF 512
Query: 369 LLVSLEADHTC--CRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGI 422
+ +L+ADHTC R+ ++QA ++AR IR P + ++++ R + G+
Sbjct: 513 TIRTLQADHTCEGVRNLHHQQASVGWVARSVESRIRDNPQVKPREILQDIRDQHGV 680
>AL375901 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 493
Score = 50.4 bits (119), Expect = 3e-06
Identities = 25/137 (18%), Positives = 63/137 (45%)
Frame = +3
Query: 618 HRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPP 677
H C++++ NFR + ++ W A A T E+ + + M E+++ + PP
Sbjct: 24 HGFCLRYVSENFRDTFKNTKLVNIFWNAVYALTAAEFESKITEMIEVSQDVISWFQHFPP 203
Query: 678 KMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQLML 737
+W AY + + E LE + P++ ++E ++ + + ++++ +
Sbjct: 204 FLWA-VAYFDGVRYGHFTLGVTELLYNWALECHELPVVQMMEYIRQQMTSWFNDRREVGM 380
Query: 738 RWRTALCPMIQQKLEDS 754
W + L P ++++ ++
Sbjct: 381 EWTSILVPSAEKRISEA 431
>AL375902 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 542
Score = 41.2 bits (95), Expect = 0.002
Identities = 15/23 (65%), Positives = 18/23 (78%)
Frame = +2
Query: 789 SRSCACRRWDLSGIPCAHAVASM 811
SR C+CRRW L G PCAHA A++
Sbjct: 5 SRECSCRRWQLYGYPCAHAAAAL 73
>TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|AI994480
gb|T44186 gb|Z30806 come from this gene. {Arabidopsis
thaliana}, partial (37%)
Length = 699
Score = 38.5 bits (88), Expect = 0.013
Identities = 48/163 (29%), Positives = 63/163 (38%)
Frame = +3
Query: 127 PVNDAPNSPEVVHEVPNSAPVNDALNSSSALVHEVPNSAPVNDALNSSSALVHEVPNSAP 186
P P+ E H +PN P L S++ + E VN +SS A VPN
Sbjct: 93 PTKRKPDQEET-HHLPNKNP---KLTSTTTVDQETELPTTVN---SSSDA----VPN--- 230
Query: 187 VNEPLNESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPP 246
NEP N ++D+ +D + EEN D V+ DR
Sbjct: 231 -NEPENNTDDDD-----------------EDDEDEENSDEEPVV-----------DRKGK 323
Query: 247 FVQGSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEG 289
+ D G G IEE EDED D SD SDD+ +G
Sbjct: 324 GIMRD-DKGKGKMIEE-------EDEDDDDSDDSDDSDDDSDG 428
>BE239532
Length = 645
Score = 34.3 bits (77), Expect = 0.24
Identities = 17/50 (34%), Positives = 31/50 (62%), Gaps = 6/50 (12%)
Frame = +3
Query: 778 RDNEKYSVNLESRS----CACRRWDLSGIPCAHA--VASMWFGRRVPEDY 821
+ N+KY+V +S + C+CR++D GI C+HA + ++ R+P+ Y
Sbjct: 186 KPNKKYAVTFDSSNMKINCSCRKFDSMGILCSHALRIYNIKGILRIPDQY 335
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 32.7 bits (73), Expect = 0.71
Identities = 28/111 (25%), Positives = 46/111 (41%)
Frame = +1
Query: 188 NEPLNESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPF 247
N E++D S D+ E + +DV+ +D D +E++ E D D
Sbjct: 271 NHTCEENKDGSETEDDDD-EDDDDDVNDEDDDNDEDFSGDE------------DDEDADP 411
Query: 248 VQGSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFK 298
V G G ++ D +D+D D+ D + +DEEE + P K
Sbjct: 412 EDDPVPNGAGGSDDDD---EDDDDDDDDNDDGEDEDEDEEEDDDEDQPPSK 555
>TC78566 similar to GP|20466388|gb|AAM20511.1 putative chloroplast outer
membrane protein {Arabidopsis thaliana}, partial (46%)
Length = 2245
Score = 32.7 bits (73), Expect = 0.71
Identities = 23/75 (30%), Positives = 32/75 (42%)
Frame = +3
Query: 269 FEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDYA 328
F DED + DLD PSD +E LP FK + + + D + K
Sbjct: 615 FSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQIRNLSRAQKKAYLDEVEYREK--- 785
Query: 329 LQQKKDLTFKKDDKK 343
L KK L ++K +K
Sbjct: 786 LFMKKQLKYEKKQRK 830
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 32.7 bits (73), Expect = 0.71
Identities = 51/220 (23%), Positives = 82/220 (37%), Gaps = 8/220 (3%)
Frame = +3
Query: 94 VYVEHDSEPEKARESDGDVEIVERSVNDAPNSAPVNDAPNS-PEVVHEVPNSAPVNDALN 152
V + +S E++ ES+ + +V+ A + + PE E +S +
Sbjct: 273 VVKKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDS-------D 431
Query: 153 SSSALVHEVPNSAPVNDALNSSSALVHEVPNSAPVNEPLNESEDESYIPG-----SDEYE 207
SSS+ EV PV+ A+ S + E ES DE P S
Sbjct: 432 SSSSDEEEVKK--PVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSKNGS 605
Query: 208 GESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNGARIEEPATFS 267
++ + D+ D +E+ D E + A + P GSV PA +
Sbjct: 606 APAKKAASDEEDTDESSD-------EDEEDEKPAAKAVPSKNGSV----------PAKKA 734
Query: 268 DFE--DEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEE 305
D E DED +SSD + + ++ P KA +E
Sbjct: 735 DTESSDEDSESSDEEDKKPAAKASKNVSAPTKKAASSSDE 854
>TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidopsis
thaliana}, partial (6%)
Length = 890
Score = 32.7 bits (73), Expect = 0.71
Identities = 24/73 (32%), Positives = 33/73 (44%), Gaps = 12/73 (16%)
Frame = +1
Query: 230 LPAESLINIATADRDPPFVQ--GSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSD--- 284
+P E + T DPP V+ G+ D +G+ E + S+ E E SS S SD
Sbjct: 331 IPIEKSVERITLTGDPPSVEVNGNEDKDDGSEGESESESSESESESSTSSSSTSGSDSDE 510
Query: 285 -------DEEEGR 290
DEEEG+
Sbjct: 511 DDEDDAEDEEEGQ 549
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 32.7 bits (73), Expect = 0.71
Identities = 27/132 (20%), Positives = 52/132 (38%)
Frame = +3
Query: 157 LVHEVPNSAPVNDALNSSSALVHEVPNSAPVNEPLNESEDESYIPGSDEYEGESEDVSVD 216
L++ + AP A++S L H + E +++ED+ D+ + E +D +
Sbjct: 210 LINAANSVAPKLTAMDSRFPLEHLLGRKPAYKENKSDTEDDEDDDDDDDVQDEDDDGEEE 389
Query: 217 DSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNGARIEEPATFSDFEDEDGDS 276
D +E + + D P G+ +G + +D+DGD
Sbjct: 390 DYSGDEGEEEGD-------------PEDDPEANGAGGSDDG----------EDDDDDGDE 500
Query: 277 SDLDSPSDDEEE 288
D + D+E+E
Sbjct: 501 EDEEDGEDEEDE 536
>TC80072 similar to GP|14423520|gb|AAK62442.1 Unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 746
Score = 32.3 bits (72), Expect = 0.92
Identities = 25/103 (24%), Positives = 43/103 (41%), Gaps = 3/103 (2%)
Frame = +1
Query: 193 ESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRD---PPFVQ 249
+ +D + D Y +ED+ + + + D ++P + + A +D D P F
Sbjct: 301 KKKDATTTSRRDRYNFSAEDMDDEIDIFHKQRD---IVPLDINADSAESDEDDEMPIFND 471
Query: 250 GSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSK 292
+D +D DEDGD + D DDE +G+ K
Sbjct: 472 KDID-------------NDETDEDGDEEEDDDDEDDEYDGKEK 561
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 32.3 bits (72), Expect = 0.92
Identities = 26/106 (24%), Positives = 47/106 (43%), Gaps = 5/106 (4%)
Frame = +1
Query: 609 LAELVGDVEHRLCIKHLYGNFR---KKYPGAE-MKQALWRAARASTLPEWYAAMEVMKEL 664
+ ++ D HRLC HL+ N + KK P E ++A++ ++++ + EL
Sbjct: 229 IKQVFPDASHRLCAWHLHKNAQENIKKTPFLEGFRKAMYSNFTPEQFEDFWSELIQKNEL 408
Query: 665 NEAAFQDMMNLPPKMWTRSAYSRDTQCD-LQVNNMCEAFNRAILEY 709
A+ +W +AY RD ++ + CEA N + Y
Sbjct: 409 EGNAWVIKTYANKSLWA-TAYLRDKFFGRIRTTSQCEAINAIVKTY 543
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 32.0 bits (71), Expect = 1.2
Identities = 35/119 (29%), Positives = 45/119 (37%), Gaps = 5/119 (4%)
Frame = +3
Query: 192 NESEDESYIPGSDEY-EGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQG 250
N+ EDE G + EGE E S D Y N + N + + + P G
Sbjct: 429 NDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSN-----------NKSNSKKAPEGGAG 575
Query: 251 SVDVGNGARIEEPATFSDFEDEDGD----SSDLDSPSDDEEEGRSKRLPRFKAVQDGEE 305
D NG + +DEDGD D D DDEEEG + +D EE
Sbjct: 576 GADE-NG---------EEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEE 722
>BF005355 similar to GP|9758230|dbj| emb|CAB61996.1~gene_id:MZF18.6~strong
similarity to unknown protein {Arabidopsis thaliana},
partial (8%)
Length = 582
Score = 30.8 bits (68), Expect = 2.7
Identities = 16/47 (34%), Positives = 27/47 (57%)
Frame = +1
Query: 183 NSAPVNEPLNESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEV 229
+ + V + +NE+EDE+ S+ S+D +DD D E DWA++
Sbjct: 214 DDSDVEDDVNENEDENEDYDSEWLAVLSDDDELDDDDDEALADWAKL 354
>TC92765 similar to EGAD|119550|127787 hypothetical protein F49E11.11
{Caenorhabditis elegans}, partial (11%)
Length = 972
Score = 30.8 bits (68), Expect = 2.7
Identities = 18/66 (27%), Positives = 30/66 (45%)
Frame = +1
Query: 261 EEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKDLF 320
EE +D EDE+ D D+D+ + + R + + ++ DG + VG D
Sbjct: 574 EEVGNVTDLEDEENDPVDIDNYRVESKSSRVEVVQIPYSLTDGVAIVGLVGDVVDGNDEG 753
Query: 321 KQAVKD 326
KQ +D
Sbjct: 754 KQPTQD 771
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,524,659
Number of Sequences: 36976
Number of extensions: 434493
Number of successful extensions: 3002
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 2167
Number of HSP's successfully gapped in prelim test: 104
Number of HSP's that attempted gapping in prelim test: 649
Number of HSP's gapped (non-prelim): 2432
length of query: 994
length of database: 9,014,727
effective HSP length: 106
effective length of query: 888
effective length of database: 5,095,271
effective search space: 4524600648
effective search space used: 4524600648
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0112.15


