

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0112.11
(1331 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
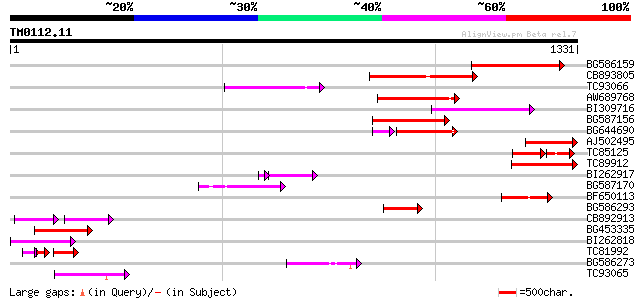
Sequences producing significant alignments: (bits) Value
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 304 1e-82
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 273 3e-73
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 187 3e-47
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 174 2e-43
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 166 4e-41
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 166 7e-41
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 141 2e-40
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 149 5e-36
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 86 1e-30
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 122 1e-27
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 108 3e-25
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 112 1e-24
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 103 4e-22
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 102 8e-22
CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabi... 73 3e-21
BG453335 weakly similar to GP|18071369|g putative gag-pol polypr... 98 2e-20
BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 93 6e-19
TC81992 weakly similar to PIR|T47841|T47841 hypothetical protein... 64 1e-18
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 90 7e-18
TC93065 84 3e-16
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 304 bits (779), Expect = 1e-82
Identities = 143/218 (65%), Positives = 179/218 (81%)
Frame = +1
Query: 1085 VEILQNSEGIFMCQSKYAKEVLERFGMKNSNPVRNPVVPGTKLNKRGCGSEVDATQYKQM 1144
VE++QN EGI++CQ KY ++LERFGM+ SN RNP+ P KL K G +VDAT+YKQ+
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1145 IGSLMYLTVSRPDLMYSVSLVSRYMEKPTELHVQIVKRVMRYLNGTVDMGIQYRRSGSMK 1204
+G LMYL +RPDLMY +SL+SR+M PTELH+ VKRV+RYLNGT+++GI Y+R+GS K
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1205 LISYTDSDYAGDVDDRKSTSGYVFMLGSGSIAWSSKKQAVVSLSTTEAEFIAAVSCACQC 1264
L +YTDSDYAGD+DDRKSTSGYVFML SG+++WSSKKQ VV+LSTT+AEFIAA CACQ
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 1265 IWLRRILFSLGQVQGKCSTIYCDNSSTIKLSKNPVMHG 1302
+W+RR+L LG Q T+YCDN+STIKLSKNPV+HG
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLHG 654
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 273 bits (698), Expect = 3e-73
Identities = 136/257 (52%), Positives = 190/257 (73%), Gaps = 5/257 (1%)
Frame = +3
Query: 846 VSIHSDPMTYDEAAKSDHWRKAMDLEIQAIERNNTWQLAELPKGDKSIGVKWVYKTKLNE 905
+++ SDP T++EA KS+ WR +M+ E++A ERNNTW+L +L G K+IG+KW++KTKLNE
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 906 KGEIEKHKARLVAKGYAQQQGIDFTEVFAPVARWDTIRTILAIAAV---RGWNVFQLDVK 962
GEIEK+KARLVAKGY+QQ G+D+TEVFAPVARWDTIR ++A+AA G + +
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KA 383
Query: 963 SAFLHGKIDETVFVDQPQGYQVRGAERKVY--KLHKALYGLRQAPRAWYSRIDSYFQKQG 1020
+ + + + + ++ R R+V ++ +ALYGL+QAPRAWYSRI++YF K+G
Sbjct: 384 HSCMEN*MRKFLLINH------RVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEG 545
Query: 1021 FVRSDSDHTLYVKQGENGLAIIVSLYVDDLIYTGNCELLISEFKASMEREFDMSDLGKMS 1080
F + +HTL+VK E G +I+SLYVDDLI+ GN E + EFK SM++EF+MSDLGKM
Sbjct: 546 FEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMH 725
Query: 1081 YFLGVEILQNSEGIFMC 1097
YFLGVE+ QN +GI++C
Sbjct: 726 YFLGVEVTQNEKGIYIC 776
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 187 bits (474), Expect = 3e-47
Identities = 105/240 (43%), Positives = 145/240 (59%), Gaps = 4/240 (1%)
Frame = +1
Query: 504 STWRAERKLQLIHSDICGPIKPKSNSQKRYFITFIDDFCRKTWVYLLSEKSGALESFKKF 563
+T R + L IHSD+ GP K S +RY +T IDDF RK WVY L K+ +FKK+
Sbjct: 73 ATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKW 252
Query: 564 KLLAEKESGEKLSCLRTDRGGEFNSREFTEFCELNGIKRQLTASYTPQQNGVSERKNRTI 623
++L E ++G+ + L TD EF S +F EFC +GI R T PQQNGV+ER RT+
Sbjct: 253 RILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTL 432
Query: 624 MNMVRCMLTEKKV--PREFWPEAVNWSVYIQNRSPTIAVKNITPEECWSGKKPSVNFFRI 681
+ RCML+ + R+ W EA + + ++ NRSP A+ PE+ WSG + RI
Sbjct: 433 LERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRI 612
Query: 682 FGCVAYAHVPDSQRRKLDNRSIKCIYLGSSEESKAYRLY--DPVSKKIIINRDVVFDESS 739
FGC AYA V D KL R+ +CI+L + ESK YRL+ DP S+K+I++RDV F+E +
Sbjct: 613 FGCPAYALVNDG---KLAPRAGECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDA 783
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 174 bits (442), Expect = 2e-43
Identities = 88/192 (45%), Positives = 124/192 (63%)
Frame = +1
Query: 864 WRKAMDLEIQAIERNNTWQLAELPKGDKSIGVKWVYKTKLNEKGEIEKHKARLVAKGYAQ 923
W +AM E +A+ N TW L LP K+IG KWVY+ K N G + K KARLVAKG++Q
Sbjct: 97 WLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQ 276
Query: 924 QQGIDFTEVFAPVARWDTIRTILAIAAVRGWNVFQLDVKSAFLHGKIDETVFVDQPQGYQ 983
G D+TE F+PV + TIR IL IA W + Q+D+ +AFL+G + E V++ QPQG++
Sbjct: 277 TLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFE 456
Query: 984 VRGAERKVYKLHKALYGLRQAPRAWYSRIDSYFQKQGFVRSDSDHTLYVKQGENGLAIIV 1043
+ V KL+K+LYGL+QAPRAWY + S + GF +S D +L + +NG I +
Sbjct: 457 AAN-KSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLI-YNQNGACIYL 630
Query: 1044 SLYVDDLIYTGN 1055
+YVDD++ TG+
Sbjct: 631 XIYVDDILITGS 666
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 166 bits (421), Expect = 4e-41
Identities = 88/243 (36%), Positives = 140/243 (57%)
Frame = +2
Query: 990 KVYKLHKALYGLRQAPRAWYSRIDSYFQKQGFVRSDSDHTLYVKQGENGLAIIVSLYVDD 1049
KV +L K++YGL+QA R WYS++ G+++S SD +L+ K ++ ++ +YVDD
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLL-VYVDD 193
Query: 1050 LIYTGNCELLISEFKASMEREFDMSDLGKMSYFLGVEILQNSEGIFMCQSKYAKEVLERF 1109
++ GN I K + F + DLG + YFLG+E+ ++ +GI + Q KY E+LE
Sbjct: 194 IVLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDS 373
Query: 1110 GMKNSNPVRNPVVPGTKLNKRGCGSEVDATQYKQMIGSLMYLTVSRPDLMYSVSLVSRYM 1169
G P KL+ D TQY+++IG L+YLT +RPD+ ++V +S+++
Sbjct: 374 GNLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFV 553
Query: 1170 EKPTELHVQIVKRVMRYLNGTVDMGIQYRRSGSMKLISYTDSDYAGDVDDRKSTSGYVFM 1229
KP ++H Q RV++YL G+ Y + ++KL S+ DSD+A RKS +GY
Sbjct: 554 SKPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVF 733
Query: 1230 LGS 1232
LGS
Sbjct: 734 LGS 742
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 166 bits (419), Expect = 7e-41
Identities = 73/180 (40%), Positives = 120/180 (66%)
Frame = -1
Query: 852 PMTYDEAAKSDHWRKAMDLEIQAIERNNTWQLAELPKGDKSIGVKWVYKTKLNEKGEIEK 911
P +Y+EA + W++++ E A+ +N+TW +ELPKG K++ +W++ K G IE+
Sbjct: 558 PRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIER 379
Query: 912 HKARLVAKGYAQQQGIDFTEVFAPVARWDTIRTILAIAAVRGWNVFQLDVKSAFLHGKID 971
K RLVA+G+ G D+ E FAPVA+ TIR +L++A GW ++Q+DVK+AFL G+++
Sbjct: 378 KKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELE 199
Query: 972 ETVFVDQPQGYQVRGAERKVYKLHKALYGLRQAPRAWYSRIDSYFQKQGFVRSDSDHTLY 1031
+ V++ P G + V +L KA+YGL+Q+PRAWY+++ + +GF +S+ DHTL+
Sbjct: 198 DEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 141 bits (355), Expect(2) = 2e-40
Identities = 66/142 (46%), Positives = 101/142 (70%)
Frame = -2
Query: 909 IEKHKARLVAKGYAQQQGIDFTEVFAPVARWDTIRTILAIAAVRGWNVFQLDVKSAFLHG 968
I ++K++LV +GY Q++GID+ E F+PVAR + IR ++A AA G+ ++Q+DVKSAF++G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 969 KIDETVFVDQPQGYQVRGAERKVYKLHKALYGLRQAPRAWYSRIDSYFQKQGFVRSDSDH 1028
+ E VFV QP G++ V++L+K LYGL+QAPRAWY R+ + K GF R D+
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 1029 TLYVKQGENGLAIIVSLYVDDL 1050
TL++ + E L +I+ +YVDD+
Sbjct: 64 TLFLLKRE*EL-LIIQVYVDDI 2
Score = 44.3 bits (103), Expect(2) = 2e-40
Identities = 18/53 (33%), Positives = 27/53 (49%)
Frame = -3
Query: 851 DPMTYDEAAKSDHWRKAMDLEIQAIERNNTWQLAELPKGDKSIGVKWVYKTKL 903
+P EA + W +M E+ ER+ W L P+G IG +WV++ KL
Sbjct: 588 EPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNKL 430
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 149 bits (377), Expect = 5e-36
Identities = 69/121 (57%), Positives = 91/121 (75%)
Frame = +2
Query: 1211 SDYAGDVDDRKSTSGYVFMLGSGSIAWSSKKQAVVSLSTTEAEFIAAVSCACQCIWLRRI 1270
SD+AGD + RKSTSGY F LG+G+I+WSSKKQ VV+ ST EAE+IA+ SCA Q +WLRRI
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1271 LFSLGQVQGKCSTIYCDNSSTIKLSKNPVMHGRSKHINVRYHFLRDLVRDGRIELVHCST 1330
L + Q + IYCDN S I LSKNPV HGRSKHI++++H +R+L+ + + + +C T
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1331 E 1331
E
Sbjct: 362 E 364
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 85.5 bits (210), Expect(2) = 1e-30
Identities = 42/78 (53%), Positives = 57/78 (72%)
Frame = +1
Query: 1180 VKRVMRYLNGTVDMGIQYRRSGSMKLISYTDSDYAGDVDDRKSTSGYVFMLGSGSIAWSS 1239
VKR+MRY+ GT + + + S + + Y DSD+AGD D RKST+GYVF L G+++W S
Sbjct: 1 VKRIMRYIKGTSGVAVCFGGS-ELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 1240 KKQAVVSLSTTEAEFIAA 1257
K Q VV+LSTTEAE++AA
Sbjct: 178 KLQTVVALSTTEAEYMAA 231
Score = 67.4 bits (163), Expect(2) = 1e-30
Identities = 26/66 (39%), Positives = 48/66 (72%), Gaps = 1/66 (1%)
Frame = +3
Query: 1261 AC-QCIWLRRILFSLGQVQGKCSTIYCDNSSTIKLSKNPVMHGRSKHINVRYHFLRDLVR 1319
AC + IW++R++ LG Q + T+YCD+ S + +++NP H R+KHI ++YHF+R++V
Sbjct: 240 ACKEAIWMQRLMEELGHKQEQI-TVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVE 416
Query: 1320 DGRIEL 1325
+G +++
Sbjct: 417 EGSVDM 434
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 122 bits (305), Expect = 1e-27
Identities = 63/156 (40%), Positives = 101/156 (64%), Gaps = 2/156 (1%)
Frame = +1
Query: 1178 QIVKRVMRYLNGTVDMGIQYRRSGSMK--LISYTDSDYAGDVDDRKSTSGYVFMLGSGSI 1235
Q +K V++YLN ++ ++Y ++ + L Y D+DYAG+VD RKS SG+VF L +I
Sbjct: 1 QALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTI 180
Query: 1236 AWSSKKQAVVSLSTTEAEFIAAVSCACQCIWLRRILFSLGQVQGKCSTIYCDNSSTIKLS 1295
+W + +Q+VV+LSTT+AE+IA V IWL+ ++ LG Q + I+CD+ S I L+
Sbjct: 181 SWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQ-EYVKIHCDSQSAIHLA 357
Query: 1296 KNPVMHGRSKHINVRYHFLRDLVRDGRIELVHCSTE 1331
+ V H R+KHI++R HF+RD++ I + ++E
Sbjct: 358 NHQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASE 465
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 108 bits (270), Expect(2) = 3e-25
Identities = 50/116 (43%), Positives = 69/116 (59%)
Frame = +1
Query: 607 SYTPQQNGVSERKNRTIMNMVRCMLTEKKVPREFWPEAVNWSVYIQNRSPTIAVKNITPE 666
++TPQQNGV+ER NRT++ R ML + + FW EAV + Y+ NRSP+ + TP
Sbjct: 79 AHTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPM 258
Query: 667 ECWSGKKPSVNFFRIFGCVAYAHVPDSQRRKLDNRSIKCIYLGSSEESKAYRLYDP 722
E W GK + +FGC Y +R KLD +S KCI+LG ++ K Y L+DP
Sbjct: 259 EMWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWDP 426
Score = 26.2 bits (56), Expect(2) = 3e-25
Identities = 13/27 (48%), Positives = 16/27 (59%), Gaps = 1/27 (3%)
Frame = +3
Query: 584 GEFNSREFTEFCELNGIKRQLT-ASYT 609
GE+ EF FC+ GI RQ T +YT
Sbjct: 9 GEYVDGEFLAFCKQEGIVRQFTGCTYT 89
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 112 bits (280), Expect = 1e-24
Identities = 66/205 (32%), Positives = 106/205 (51%)
Frame = -3
Query: 443 STTIKDESELWHKRYGHLGYSGLKILAQKAMVKGLPMLGELDKPCEECLKGKQHIDPFPQ 502
S++ ++ LWH R GH L ++ LP + +K CE C+ GK + FP+
Sbjct: 611 SSSSLNKDALWHARLGHPHGRALNLM--------LPGVVFENKNCEACILGKHCKNVFPR 456
Query: 503 KSTWRAERKLQLIHSDICGPIKPKSNSQKRYFITFIDDFCRKTWVYLLSEKSGALESFKK 562
ST E LI++D+ S +YF+TFID+ + TW+ L+ K +++FK
Sbjct: 455 TSTVY-ENCFDLIYTDLW-TAPSLSRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKN 282
Query: 563 FKLLAEKESGEKLSCLRTDRGGEFNSREFTEFCELNGIKRQLTASYTPQQNGVSERKNRT 622
F+ K+ LR+D GGE+ S F + +GI Q + YTPQQNGV++RKN+
Sbjct: 281 FQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKH 102
Query: 623 IMNMVRCMLTEKKVPREFWPEAVNW 647
+M + R ++ + V + +NW
Sbjct: 101 LMEVARSLMFQANVSTACY--LINW 33
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 103 bits (257), Expect = 4e-22
Identities = 50/123 (40%), Positives = 79/123 (63%), Gaps = 3/123 (2%)
Frame = +1
Query: 1155 RPDLMYSVSLVSRYMEKPTELHVQIVKRVMRYLNGTVDMGIQY---RRSGSMKLISYTDS 1211
RPD+ YSVS++S++M P + H+ R++RY+ GT++ G+ + +S +LI Y+DS
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 1212 DYAGDVDDRKSTSGYVFMLGSGSIAWSSKKQAVVSLSTTEAEFIAAVSCACQCIWLRRIL 1271
D+ GD R+STSGYVF +I+W +KKQ + +LS+ EAE+IA Q +WL ++
Sbjct: 301 DWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 1272 FSL 1274
L
Sbjct: 472 KEL 480
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 102 bits (255), Expect = 8e-22
Identities = 51/92 (55%), Positives = 66/92 (71%)
Frame = +2
Query: 877 RNNTWQLAELPKGDKSIGVKWVYKTKLNEKGEIEKHKARLVAKGYAQQQGIDFTEVFAPV 936
+ T +L + P G K IG++W+YK K NE G + K+KARLVAKGY +QQGIDF EVFAPV
Sbjct: 47 QKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPV 226
Query: 937 ARWDTIRTILAIAAVRGWNVFQLDVKSAFLHG 968
R +TI +LA+AA G + +DVK AFL+G
Sbjct: 227 VRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabidopsis
thaliana}, partial (35%)
Length = 837
Score = 73.2 bits (178), Expect(2) = 3e-21
Identities = 43/118 (36%), Positives = 68/118 (57%), Gaps = 1/118 (0%)
Frame = +1
Query: 128 ETVVNYFARTLSIVNKMKIQGESIKETLIVEKILRSLTEKFNYVVCSIEESNDVDTITID 187
E V YF+R +SI N++K E +++ I+EKILRSL KF ++V IEE+ D++T+TI+
Sbjct: 472 ERVRVYFSRVISISNQLKRNSEKLEDVRIMEKILRSLDPKFEHIVEKIEETKDLETMTIE 651
Query: 188 QLQSSLLVQEQRMKGYRE-EEQALRATSSGKSERGRGRTGNRGRGRGRGSFNKESVEC 244
+LQ SL E++ K ++ E + K E R + +GRG N + + C
Sbjct: 652 KLQGSLQAYEEKHKKKKDITE*LFKMQLKEKDEGQRNERIQQSQGRGST*KNCKFINC 825
Score = 48.5 bits (114), Expect(2) = 3e-21
Identities = 30/105 (28%), Positives = 53/105 (49%), Gaps = 3/105 (2%)
Frame = +3
Query: 12 VPKFDGH-YDHWALLMENLLRSKEYWPIVADGVPALAP--NANAEQIKIHEEAKLKDLKA 68
VP G YD+ + M LL + + W +V G + Q ++++ +D KA
Sbjct: 126 VPLLKGSTYDNCFIKMMALLGAHDVWEVVEKGYKESQDEDSLTKAQRDTLKDSRKRDKKA 305
Query: 69 KNYLFQSIERSILETILNMETSKEIWDSMRLKNQGSTKVKRVQLQ 113
++Q+++ E I N ++KE W+ ++ QG KVK+V+LQ
Sbjct: 306 LFLIYQALDEDEFEKISNATSAKEAWEKLKTSCQGEDKVKKVRLQ 440
>BG453335 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (11%)
Length = 660
Score = 97.8 bits (242), Expect = 2e-20
Identities = 47/136 (34%), Positives = 91/136 (66%)
Frame = +1
Query: 58 HEEAKLKDLKAKNYLFQSIERSILETILNMETSKEIWDSMRLKNQGSTKVKRVQLQALRR 117
H+E K KD K + S++ + I SKE + + ++G KV++++LQ+LRR
Sbjct: 109 HKELKKKDCKELFLIQ*SLDEGNFKRISKSTRSKEASNILEKYHEGDDKVEQIKLQSLRR 288
Query: 118 EFEVLGMKEGETVVNYFARTLSIVNKMKIQGESIKETLIVEKILRSLTEKFNYVVCSIEE 177
+F+++ M++ + + +Y ++ +++VN++K GE + + IVEKI+R+L+ +F+++V +I E
Sbjct: 289 KFKLMQMEDEKKIADYISKLINVVNQIKAYGEVVTDQQIVEKIMRTLSPRFDFIVVAIHE 468
Query: 178 SNDVDTITIDQLQSSL 193
S DV T+ I++L+SSL
Sbjct: 469 SKDVKTLKIEELKSSL 516
>BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (24%)
Length = 615
Score = 93.2 bits (230), Expect = 6e-19
Identities = 49/157 (31%), Positives = 88/157 (55%), Gaps = 3/157 (1%)
Frame = +1
Query: 1 MAESSNFSQPAVPKFDG-HYDHWALLMENLLRSKEYWPIVADG--VPALAPNANAEQIKI 57
M +++ P +P FDG +Y WA +E L + + W V + V L+ N QIK
Sbjct: 142 MDSETSYKAPPLPVFDGENYHIWAARIEAYLEANDLWEAVEEDYEVLPLSDNPTMAQIKN 321
Query: 58 HEEAKLKDLKAKNYLFQSIERSILETILNMETSKEIWDSMRLKNQGSTKVKRVQLQALRR 117
H+E K + KA+ LF ++ I I+ ++++ EIW+ ++ + +G +++ +Q L R
Sbjct: 322 HKERKTRKSKARATLFAAVSEEIFTRIMTIKSAFEIWNFLKTEYEGDERIRGMQALNLIR 501
Query: 118 EFEVLGMKEGETVVNYFARTLSIVNKMKIQGESIKET 154
EFE+ MKE ET+ Y + +SI NK+++ G + ++
Sbjct: 502 EFEMQKMKESETIKEYANKLISIANKVRLLGSELXDS 612
>TC81992 weakly similar to PIR|T47841|T47841 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (3%)
Length = 952
Score = 63.9 bits (154), Expect(3) = 1e-18
Identities = 29/60 (48%), Positives = 46/60 (76%)
Frame = +3
Query: 102 QGSTKVKRVQLQALRREFEVLGMKEGETVVNYFARTLSIVNKMKIQGESIKETLIVEKIL 161
Q + + KR QLQAL R++ +L MK GE++++YFART++IVNK+++ GE + ++EKIL
Sbjct: 720 QVTARGKRAQLQAL*RDW*ILQMKNGESIIDYFARTVTIVNKIRMHGEQMSNLTVIEKIL 899
Score = 43.1 bits (100), Expect(3) = 1e-18
Identities = 21/30 (70%), Positives = 25/30 (83%)
Frame = +2
Query: 64 KDLKAKNYLFQSIERSILETILNMETSKEI 93
K KAKN+LFQ+I+R +LETIL ETSKEI
Sbjct: 608 KIFKAKNHLFQAIDRVVLETILKNETSKEI 697
Score = 25.4 bits (54), Expect(3) = 1e-18
Identities = 15/42 (35%), Positives = 21/42 (49%), Gaps = 2/42 (4%)
Frame = +1
Query: 30 LRSKEYWPIVADGVP--ALAPNANAEQIKIHEEAKLKDLKAK 69
L SKEYW ++ G+P A+ + K +E KL D K
Sbjct: 499 LASKEYWNLIYPGIPQQAVGVVLTENEKKTLDELKL*DF*GK 624
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 89.7 bits (221), Expect = 7e-18
Identities = 58/183 (31%), Positives = 93/183 (50%), Gaps = 7/183 (3%)
Frame = -2
Query: 650 YIQNRSPTIAVKNITPEECWSGKKPSVNFFRIFGCVAYAHVPDSQRRKLDNRSIKCIYLG 709
Y+ NR PT +K+ P E + +KPS+ + R+FGC+ Y VP R KL+ RS K +++G
Sbjct: 698 YLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMFIG 519
Query: 710 SSEESKAYRLYDPVSKKIIINRDVVFDESSGWDWSSGAEQMEFFDEMVDTNEVSTVPILD 769
S K Y+ YDP +++++++RDV F E G+ E + D D V V IL+
Sbjct: 518 YSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLR--DLTSDKAGVLRV-ILE 348
Query: 770 EVEHQVHSE--TNDTSPELEDNGEASVSASP-----GVAEEERERRGRRNVQQPVWMKEF 822
+ +++ + T PE N + +P G +E E + G+ V +EF
Sbjct: 347 GLGIKMNQDQSTRSRQPEESSNEPRRAAQTPHLDHEGGSEPETQENGQEGVGLSEEGEEF 168
Query: 823 VSG 825
SG
Sbjct: 167 NSG 159
>TC93065
Length = 783
Score = 84.3 bits (207), Expect = 3e-16
Identities = 60/190 (31%), Positives = 94/190 (48%), Gaps = 15/190 (7%)
Frame = +2
Query: 106 KVKRVQLQALRREFEVLGMKEGETVVNYFARTLSIVNKMKIQGESIKETLIVEKILRSLT 165
K K + LRREFE L MKE ETV + R +V ++++ GE + + +VEKIL L
Sbjct: 188 KNKENESAKLRREFEALKMKETETVREFSDRISKVVTQIRLLGEDLSDQRVVEKILVCLP 367
Query: 166 EKFNYVVCSIEESNDVDTITIDQLQSSLLVQEQRMKGYREE--EQALRATSSGKSE--RG 221
E F + S+EE+ + IT+ +L ++L EQR EE E A A + GK++ +
Sbjct: 368 EMFEAKISSLEENKNFSEITVAELVNALQASEQRRSLRMEENVEGAFLANNKGKNQSFKS 547
Query: 222 RGRTG-------NRGRGRGRGSFNKESVECYKCHKLGHFQYECPT----WEESANYAELH 270
G+ + + + + V+C C++LGH + C E+ A E H
Sbjct: 548 FGKKKFPPCPHCKKDTHLDKFCWYRPGVKCRACNQLGHVEKVCKNKTNQQEQEARVVEHH 727
Query: 271 EDHEELLLMA 280
++ EE L A
Sbjct: 728 QEDEEPLFKA 757
Score = 42.0 bits (97), Expect = 0.002
Identities = 24/100 (24%), Positives = 49/100 (49%)
Frame = +3
Query: 44 PALAPNANAEQIKIHEEAKLKDLKAKNYLFQSIERSILETILNMETSKEIWDSMRLKNQG 103
P L N QI+ H + K+ +A + ++ I ILN+E +KE W + + QG
Sbjct: 3 PPLPNNPTLAQIRNHSDEVAKEGRALAIIHAALHDDIFIKILNLEIAKEAWSKLMEEYQG 182
Query: 104 STKVKRVQLQALRREFEVLGMKEGETVVNYFARTLSIVNK 143
S + K++++ L + K+ + + ++ L +++K
Sbjct: 183 SERTKKMKVLNLEESLKH*K*KKLKLLESFLTEFLKLLHK 302
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,314,229
Number of Sequences: 36976
Number of extensions: 531895
Number of successful extensions: 3098
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 2887
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3016
length of query: 1331
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1223
effective length of database: 5,021,319
effective search space: 6141073137
effective search space used: 6141073137
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0112.11


