

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0109.6
(545 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
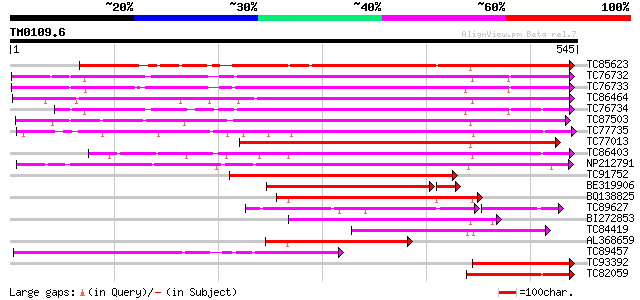
Score E
Sequences producing significant alignments: (bits) Value
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 419 e-117
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 403 e-113
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 400 e-112
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 394 e-110
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 384 e-107
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 376 e-104
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 371 e-103
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 365 e-101
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 362 e-100
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 301 6e-82
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 249 2e-66
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 217 4e-61
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 204 7e-53
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 143 2e-46
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 174 1e-43
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 173 1e-43
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 165 4e-41
TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE... 158 4e-39
TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein... 137 8e-33
TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Ar... 135 4e-32
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 419 bits (1076), Expect = e-117
Identities = 231/484 (47%), Positives = 305/484 (62%), Gaps = 8/484 (1%)
Frame = +3
Query: 68 SPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGE 127
S + +L + A+ +N S +E+ A DC EL DF+V +L+ +
Sbjct: 735 SKSDFFKVSLEIAMERAKKGEENTHAVGPKCRSPQEKAAWADCLELYDFTVQKLSQT--- 905
Query: 128 MRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQ-VTQLISN 186
QYE + WLS AL+N +TC GF D + +Y+ L+ VT+L+SN
Sbjct: 906 -----KYTKCTQYES--QTWLSTALTNLETCKNGFY--DLGVTNYVLPLLSNNVTKLLSN 1058
Query: 187 VLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVAL 246
LSL + +P++ P FP W+ GD++LL++ S+A+ VVA
Sbjct: 1059 TLSL----NKVPYQQP-----------SYKDGFPTWVKPGDRKLLQTSSAASKANVVVAK 1193
Query: 247 DGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITS 306
DGSG Y T+ A +AAPS S R YVIYVK G+Y E V++K K N+MLVGDGIG+TIIT
Sbjct: 1194 DGSGKYTTVKAATDAAPSGSGR-YVIYVKAGVYNEQVEIKAK--NVMLVGDGIGKTIITG 1364
Query: 307 NRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEG 366
+++ G TTFR+ATVA +G GFIA+D++FRNTAG NHQAVA R SD S FY+CS EG
Sbjct: 1365 SKSVGGGTTTFRSATVAATGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYKCSFEG 1544
Query: 367 FQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSP 426
+QDTLY HS RQFYREC IYGT+DFIFGN A VLQNC I+ R P P + +TVTAQGR P
Sbjct: 1545 YQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCNIFARNP-PAKTITVTAQGRTDP 1721
Query: 427 HQSTGFTIQDSFILA-------TQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGD 479
+Q+TG I +S + A + +YLGRPW++YSRTV++ T + G + P GWL W G+
Sbjct: 1722 NQNTGIIIHNSRVSAQSDLNPSSVKSYLGRPWQKYSRTVFMKTVLDGFINPAGWLPWDGN 1901
Query: 480 FALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKF 539
FAL TL+Y EY N G G+S + RV W GYH++ A+ AS FTV FI G+SW+ TGV F
Sbjct: 1902 FALDTLYYAEYANTGSGSSTSNRVTWKGYHVLTSASQASPFTVGNFIAGNSWIGNTGVPF 2081
Query: 540 TAGL 543
T+GL
Sbjct: 2082 TSGL 2093
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 403 bits (1036), Expect = e-113
Identities = 232/560 (41%), Positives = 330/560 (58%), Gaps = 18/560 (3%)
Frame = +2
Query: 2 ALVNALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNEL 61
++ ++++ L+ + SSS + I ++ + C + SCL ++ +E+
Sbjct: 74 SIPKTFWLVLSLVAIISSSALIISHLNKPISFFNLSSA-PNLCEHALDTKSCLTHV-SEV 247
Query: 62 TKIGPPSPT-----SVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDF 116
+ S T S L + L + + A+D S RE+IA+ DC+EL+D
Sbjct: 248 VQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEIALNDCEELMDL 427
Query: 117 SVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRR-LESYISG 175
S+ + S+ + + ++ + + WLS+ L+N TCL+G EG+ R +ES +
Sbjct: 428 SMDRVWDSVLTLTK-----NNIDSQHDAHTWLSSVLTNHATCLDGLEGSSRVVMESDLHD 592
Query: 176 SLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKP 235
+++ ++ ++S+ PP+ N E L+ D FP W+T D+ LL S
Sbjct: 593 LISRARSSLAVLVSVL---------PPKANDGFIDEKLNGD--FPSWVTSKDRRLLESSV 739
Query: 236 HRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLV 295
+A+ VVA DGSG ++T+ +AV +AP + RYVIYVKKG Y+EN+++ +K TN+MLV
Sbjct: 740 GDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLV 919
Query: 296 GDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSD 355
GDG+ TIIT + NF+ G TTF++ATVA G GFIA+D+ F+NTAGP HQAVALRV +D
Sbjct: 920 GDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGAD 1099
Query: 356 QSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQK 415
QS RC I+ FQDTLYAHS RQFYR+ I GT+DFIFGN A V Q K+ R P+ QK
Sbjct: 1100QSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQK 1279
Query: 416 VTVTAQGRKSPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTVYINTYMSG 466
VTAQGR+ P+Q+T +IQ ++ + TYLGRPWK+YSRTV + + + G
Sbjct: 1280NMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDG 1459
Query: 467 MVQPRGWLEWFG---DFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQ 523
+ P GW EW DF L TL+YGEY N G GA RV WPGYHI+K+AA AS FTV
Sbjct: 1460HIDPAGWAEWDAASKDF-LQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVT 1636
Query: 524 RFIHGDSWLPGTGVKFTAGL 543
+ I G+ WL TGV F GL
Sbjct: 1637QLIQGNVWLKNTGVAFIEGL 1696
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 400 bits (1029), Expect = e-112
Identities = 228/560 (40%), Positives = 325/560 (57%), Gaps = 18/560 (3%)
Frame = +2
Query: 2 ALVNALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNEL 61
+L ++++ L+ + SS S + I + + C + SCL ++ +E+
Sbjct: 86 SLPKTFWLILSLVAIIISSALISTHLKKPISFFHLTTVQNVYCEHAVDTKSCLAHV-SEV 262
Query: 62 TKIGPPSPTS-----VLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDF 116
+ + T VL + L + Q A+D + S RE+IA+ DC++L+D
Sbjct: 263 SHVPTLVTTKDQNLHVLLSLLTKSTTHIQNAMDTASVIKRRINSPREEIALSDCEQLMDL 442
Query: 117 SVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRR-LESYISG 175
S++ + W ++ ++ + + WLS+ L+N TCL+G EG+ R +E+ +
Sbjct: 443 SMNRI-WDT----MLKLTKNNIDSQQDAHTWLSSVLTNHATCLDGLEGSSRVVMENDLQD 607
Query: 176 SLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKP 235
+++ ++ L ++ P ++ E+L EFP W+T D+ LL +
Sbjct: 608 LISRARSSLAVFLVVF---------PQKDRDQFIDETLI--GEFPSWVTSKDRRLLETAV 754
Query: 236 HRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLV 295
+A+ VVA DGSG ++T+ EAV +AP + +YVIYVKKG Y+ENV++ K TN+MLV
Sbjct: 755 GDIKANVVVAQDGSGKFKTVAEAVASAPDNGKTKYVIYVKKGTYKENVEIGSKKTNVMLV 934
Query: 296 GDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSD 355
GDG+ TIIT N NF+ G TTF+++TVA G GFIA+D+ F+N AG HQAVALRV SD
Sbjct: 935 GDGMDATIITGNLNFIDGTTTFKSSTVAAVGDGFIAQDIWFQNMAGAAKHQAVALRVGSD 1114
Query: 356 QSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQK 415
QS RC I+ FQDTLYAHS RQFYR+ I GTIDFIFGN A V Q CK+ R P+ Q
Sbjct: 1115QSVINRCRIDAFQDTLYAHSNRQFYRDSVITGTIDFIFGNAAVVFQKCKLVARKPMANQN 1294
Query: 416 VTVTAQGRKSPHQSTGFTIQD---------SFILATQPTYLGRPWKQYSRTVYINTYMSG 466
TAQGR+ P Q+TG +IQ ++ + T+LGRPWK+YSRTV + +++
Sbjct: 1295NMFTAQGREDPGQNTGTSIQQCDLTPSSDLKPVVGSIKTFLGRPWKKYSRTVVMQSFLDS 1474
Query: 467 MVQPRGWLEWFG---DFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQ 523
+ P GW EW DF L TL+YGEY N GPGA A RV WPGYH++ AA AS FTV
Sbjct: 1475HIDPTGWAEWDAASKDF-LQTLYYGEYLNNGPGAGTAKRVTWPGYHVINTAAEASKFTVA 1651
Query: 524 RFIHGDSWLPGTGVKFTAGL 543
+ I G+ WL TGV FT GL
Sbjct: 1652QLIQGNVWLKNTGVAFTEGL 1711
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 394 bits (1011), Expect = e-110
Identities = 231/565 (40%), Positives = 326/565 (56%), Gaps = 24/565 (4%)
Frame = +1
Query: 3 LVNALFILMMLLILPSSSKSFSDEIPSEIQT--QDMQALIAQACMDIENQNSCLQNIHNE 60
L+ A + ++ +S+KS ++ I S + A++ AC C I +E
Sbjct: 133 LIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTLYPELCFSAISSE 312
Query: 61 LT---KIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTF-SVSNREQIAIGDCKELVDF 116
KI + V+S +L T + + K S++ RE+IA+ DC E +D
Sbjct: 313 PNITHKI--TNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKREKIALHDCLETIDE 486
Query: 117 SVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGF--EGTDRRLESYIS 174
++ EL + ++ + T Q+ +L+ +S+A++NQ TCL+GF + D+ + +
Sbjct: 487 TLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQ 666
Query: 175 GSLTQVTQLISNVLSL---YTQLHALPFRPPRNNTHKTSESLDLDSE----FPEWMTEGD 227
V + SN L++ T F L+ E +PEW++ GD
Sbjct: 667 EGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENGVGWPEWISAGD 846
Query: 228 QELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKR 287
+ LL+ +AD VVA DGSG+++T++EAV AAP S++RYVI +K G+Y+ENV++ +
Sbjct: 847 RRLLQGST--VKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPK 1020
Query: 288 KMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQA 347
K TNIM +GDG TIIT +RN + G TTF +ATVA+ G F+ARD++F+NTAGP HQA
Sbjct: 1021KKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQA 1200
Query: 348 VALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYT 407
VALRV +D SAFY C I +QDTLY H+ RQF+ C I GT+DFIFGN A V QNC I+
Sbjct: 1201VALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHA 1380
Query: 408 RAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTV 458
R P QK VTAQGR P+Q+TG IQ I AT+ PTYLGRPWK+YSRTV
Sbjct: 1381RRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTV 1560
Query: 459 YINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATAS 518
++ + +S ++ P GW EW G+FAL TL Y EY+N GPGA + RV W G+ ++ AA A
Sbjct: 1561FMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQ 1740
Query: 519 YFTVQRFIHGDSWLPGTGVKFTAGL 543
T FI G SWL TG F+ GL
Sbjct: 1741SSTPGNFIGGSSWLGSTGFPFSLGL 1815
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 384 bits (985), Expect = e-107
Identities = 223/517 (43%), Positives = 306/517 (59%), Gaps = 17/517 (3%)
Frame = +3
Query: 44 CMDIENQNSCLQNIHNELTKIGPPSPT-----SVLSAALRTTLNEAQGAIDNLTKFSTFS 98
C + NSCL ++ E+ + T S L + L + + A+D
Sbjct: 204 CEHAVDTNSCLTHVA-EVVQGSTLDNTKDHKLSTLISLLTKSTTHIRKAMDTANVIKRRI 380
Query: 99 VSNREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTC 158
S RE+ A+ C++L++ S+ + S+ + + ++ + + WLS+ L+N TC
Sbjct: 381 NSPREENALNVCEKLMNLSMERVWDSVLTLTK-----DNMDSQQDAHTWLSSVLTNHATC 545
Query: 159 LEGFEGTDRRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSE 218
L+G EGT R + + LI+ S L A+ PP+++ ESL+ D
Sbjct: 546 LDGLEGTSRAVME------NDIQDLIARARSSLAVLVAV--LPPKDHDEFIDESLNGD-- 695
Query: 219 FPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGL 278
FP W+T D+ LL S +A+ VVA DGSG ++T+ EAV +AP+ RYVIYVKKG+
Sbjct: 696 FPSWVTSKDRRLLESSVGDVKANVVVAKDGSGKFKTVAEAVASAPNKGTARYVIYVKKGI 875
Query: 279 YRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRN 338
Y+ENV++ TN+ML+GDG+ TIIT + N++ G TF+TATVA G FIA+D+ F+N
Sbjct: 876 YKENVEIASSKTNVMLLGDGMDATIITGSLNYVDGTGTFQTATVAAVGDWFIAQDIGFQN 1055
Query: 339 TAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAA 398
TAGP HQAVALRV SD+S RC I+ FQDTLYAH+ RQFYR+ I GTIDFIFG+ A
Sbjct: 1056TAGPQKHQAVALRVGSDRSVINRCKIDAFQDTLYAHTNRQFYRDSFITGTIDFIFGDAAV 1235
Query: 399 VLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQD---------SFILATQPTYLGR 449
VLQ CK+ R P+ Q VTAQGR P+Q+T +IQ ++ + TYLGR
Sbjct: 1236VLQKCKLVARKPMANQNNMVTAQGRIDPNQNTATSIQQCDVIPSTDLKPVIGSVKTYLGR 1415
Query: 450 PWKQYSRTVYINTYMSGMVQPRGWLEWFG---DFALTTLWYGEYRNYGPGASLAGRVKWP 506
PWK+YSRTV + + + + P GW EW DF L TL+YGEY N GPGA + RVKWP
Sbjct: 1416PWKKYSRTVVMQSLLGAHIDPTGWAEWDAASKDF-LQTLYYGEYMNSGPGAGTSKRVKWP 1592
Query: 507 GYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
GYHI+ + A A+ FTV + I G+ WL TGV F AGL
Sbjct: 1593GYHII-NTAEANKFTVAQLIQGNVWLKNTGVAFIAGL 1700
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 376 bits (965), Expect = e-104
Identities = 216/548 (39%), Positives = 310/548 (56%), Gaps = 14/548 (2%)
Frame = +2
Query: 6 ALFILMMLLILPSSSKSFSDEIPSEIQTQDMQAL--IAQACMDIENQNSCLQNIHNELTK 63
A+ I + IL + S P+ + ++ + C + NSC +I + L
Sbjct: 179 AVIIAAVAGILIHKHNTESSSSPNSLPNTELTPATSLKAVCESTQYPNSCFSSI-SSLPD 355
Query: 64 IGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAW 123
P + +L+ ++E +LT+FS + R + AIG C ++ S+ L
Sbjct: 356 SNTTDPEQLFKLSLKVAIDELSKL--SLTRFSEKATEPRVKKAIGVCDNVLADSLDRLND 529
Query: 124 SLGEMRRIRAGDT-SVQYEGNLEAWLSAALSNQDTCLEGFEGTD----RRLESYISGSLT 178
S+ + + G S ++E WLSAAL++ DTCL+ + R + I +
Sbjct: 530 SMSTI--VDGGKMLSPAKIRDVETWLSAALTDHDTCLDAVGEVNSTAARGVIPEIERIMR 703
Query: 179 QVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRS 238
T+ SN L++ +++ L +N H+ L EFPEW+ ++ LL + + +
Sbjct: 704 NSTEFASNSLAIVSKVIRLLSNFEVSNHHRR-----LLGEFPEWLGTAERRLLATVVNET 868
Query: 239 RADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDG 298
DAVVA DGSG Y+TI EA+ S +R+V+YVKKG+Y EN+D+ + N+M+ GDG
Sbjct: 869 VPDAVVAKDGSGQYKTIGEALKLVKKKSLQRFVVYVKKGVYVENIDLDKNTWNVMIYGDG 1048
Query: 299 IGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSA 358
+ +T+++ +RN++ G TF TAT AV GKGFIA+D+ F NTAG HQAVA+R SDQS
Sbjct: 1049MTETVVSGSRNYIDGTPTFETATFAVKGKGFIAKDIQFLNTAGASKHQAVAMRSGSDQSV 1228
Query: 359 FYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTV 418
FYRCS G+QDTLYAHS RQFYR+C+I GTIDFIFGN AAV QNCKI R P+ Q T+
Sbjct: 1229FYRCSFVGYQDTLYAHSNRQFYRDCDITGTIDFIFGNAAAVFQNCKIMPRQPMSNQFNTI 1408
Query: 419 TAQGRKSPHQSTGFTIQDSFILA------TQPTYLGRPWKQYSRTVYINTYMSGMVQPRG 472
TAQG+K P+Q++G IQ S PTYLGRPWK +S T+ + + + ++P G
Sbjct: 1409TAQGKKDPNQNSGIVIQKSTFTTLPGDNLIAPTYLGRPWKDFSTTIIMKSEIGSFLKPVG 1588
Query: 473 WLEWFGDF-ALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSW 531
W+ W + +++ Y EY+N GPGA + GRVKW GY A FTV FI G W
Sbjct: 1589WISWVANVEPPSSILYAEYQNTGPGADVPGRVKWAGYKPALGDEDAIKFTVDSFIQGPEW 1768
Query: 532 LPGTGVKF 539
LP V+F
Sbjct: 1769LPSASVQF 1792
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 371 bits (953), Expect = e-103
Identities = 231/583 (39%), Positives = 321/583 (54%), Gaps = 44/583 (7%)
Frame = +2
Query: 7 LFILM--MLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNELTKI 64
LFIL+ +LL+ S S S PS L + C S L I +
Sbjct: 65 LFILLAPLLLLAQSPPPSPSPPPPSSSSAACKTTLYPKLC------RSMLSAIRSS---- 214
Query: 65 GPPSPTSVLSAALRTTLNEAQGA----IDNLTKFSTFSVSNREQI-AIGDCKELVDFSVS 119
P P + +++ L A+ ID L + + S N E++ A+ DCK+L +V
Sbjct: 215 -PSDPYNYGKFSIKQNLKVARKLEKVFIDFLNRHQSSSSLNHEEVGALVDCKDLNSLNVD 391
Query: 120 ELAWSLGEMRRIRAGDTSVQYE--GNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSL 177
L E++ + +S E +E++LSA +N TC +G T + + ++ L
Sbjct: 392 YLESISDELKSASSSSSSSDTELVDKIESYLSAVATNHYTCYDGLVVTKSNIANALAVPL 571
Query: 178 TQVTQLISNVLSLYTQLHALPFRPPRNNTHK---TSESLDLDSEFPEWM----------- 223
TQ S L L T+ AL RN T K ++S + + +
Sbjct: 572 KDATQFYSVSLGLVTE--ALSKNMKRNKTRKHGLPNKSFKVRQPLEKLIKLLRTKYSCQK 745
Query: 224 -------TEGDQELLRSKPHRSRADAVVALD--GSGHYRTITEAVNAAPSHSNRR---YV 271
T ++ L S+ H + V + G ++ +I +A+ AAP+++ Y+
Sbjct: 746 TSSNCTSTRTERILKESESHGILLNDFVLVSPYGIANHTSIGDAIAAAPNNTKPEDGYYL 925
Query: 272 IYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIA 331
IYV++G Y E V + + NI+LVGDGI TIIT N + + GWTTF ++T AVSG+ FIA
Sbjct: 926 IYVREGYYEEYVIVPKHKNNILLVGDGINNTIITGNHSVIDGWTTFNSSTFAVSGERFIA 1105
Query: 332 RDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDF 391
D++FRNTAGP HQAVA+R ++D S FYRCS EG+QDTLY HS+RQFYR+C+IYGT+DF
Sbjct: 1106VDITFRNTAGPEKHQAVAVRNNADLSTFYRCSFEGYQDTLYVHSVRQFYRDCKIYGTVDF 1285
Query: 392 IFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQP------- 444
IFGN A V QNC IY R PLP QK VTAQGR P+Q+TG +IQ+ I A Q
Sbjct: 1286IFGNAAVVFQNCNIYARKPLPNQKNAVTAQGRTDPNQNTGISIQNCTIDAAQDLANDLNS 1465
Query: 445 --TYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGR 502
+YLGRPWK YSRTVY+ +Y+ VQP GWLEW G L T++YGE+ NYGPG+ R
Sbjct: 1466TMSYLGRPWKIYSRTVYMQSYIGDFVQPSGWLEWNGTVGLDTIFYGEFNNYGPGSVTNNR 1645
Query: 503 VKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGLTN 545
V+WPG+ ++ D A FTV F G++WLP T + +T GL N
Sbjct: 1646VQWPGHFLLND-TQAWNFTVLNFTLGNTWLPDTDIPYTEGLLN 1771
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 365 bits (937), Expect = e-101
Identities = 170/317 (53%), Positives = 236/317 (73%), Gaps = 9/317 (2%)
Frame = +2
Query: 222 WMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRE 281
W++ D++LL++ ++++ D VVA DG+G++ TI EA+ AAP+ S R+VI++K G Y E
Sbjct: 41 WVSPKDRKLLQAAVNQTKFDLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIKAGAYFE 220
Query: 282 NVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAG 341
NV++ +K TN+MLVGDGIGQT++ ++RN + GWTTF+++T AV G FIA+ ++F N+AG
Sbjct: 221 NVEVIKKKTNLMLVGDGIGQTVVKASRNVVDGWTTFQSSTFAVVGDKFIAKGITFENSAG 400
Query: 342 PVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQ 401
P HQAVA+R +D SAFY+CS +QDTLY HSLRQFYREC++YGT+DFIFGN A V Q
Sbjct: 401 PSKHQAVAVRNGADFSAFYQCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGNAAVVFQ 580
Query: 402 NCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILA---------TQPTYLGRPWK 452
NC +Y R P P QK TAQGR+ P+Q+TG +I + I A T +YLGRPWK
Sbjct: 581 NCNLYARKPDPKQKNLFTAQGREDPNQNTGISILNCKIAAAADLIPVQSTFKSYLGRPWK 760
Query: 453 QYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMK 512
+YSRTVY+N+++ ++ P GWLEW G FAL TL+YGEY+N GPG++ + RV WPGY ++
Sbjct: 761 KYSRTVYLNSFIDNLIDPPGWLEWNGTFALDTLFYGEYKNRGPGSNTSARVTWPGYKVIT 940
Query: 513 DAATASYFTVQRFIHGD 529
+A AS FTV++FI G+
Sbjct: 941 NATEASQFTVRQFIQGE 991
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 362 bits (929), Expect = e-100
Identities = 209/503 (41%), Positives = 293/503 (57%), Gaps = 35/503 (6%)
Frame = +3
Query: 76 ALRTTLNEAQGAIDNLTKF----STFSVSNREQIAIGDCKELVDFSVSELAWSLGEMRRI 131
+++ +L++++ D + K+ ST S + A+ DC+ L D + L+ S + +
Sbjct: 219 SVKKSLSQSRKFSDLINKYLQRRSTLSTTALR--ALQDCQSLSDLNFDFLSSSFQTVNKT 392
Query: 132 RAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRR--LESYISGSLTQVTQLISNVLS 189
S+Q E N++ LSA L+NQ TCL+G + T + ++ L+ T+L S L+
Sbjct: 393 TKFLPSLQGE-NIQTLLSAILTNQQTCLDGLKDTSSAWSFRNGLTIPLSNDTKLYSVSLA 569
Query: 190 LYTQLHALPFRPPRNNTH----KTSESLDLDSEFPEWMTEGDQELLRSKPHRS------- 238
+T+ P N T K S + P MT + + S R
Sbjct: 570 FFTKGWV---NPKTNKTSFPNSKHSNKGFKNGRLPLKMTSKTRAIYESVSRRKLLQSNQV 740
Query: 239 ------RADAVVALDGSGHYRTITEAVNAAPSHS---NRRYVIYVKKGLYRENVDMKRKM 289
R V+ DGSG++ TI +A+ AAP+ S + ++IYV G+Y E + + +K
Sbjct: 741 GEDVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSSDGYFLIYVTAGVYEEYITIDKKK 920
Query: 290 TNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVA 349
T +M++GDGI +TIIT N + + GWTTF + T AV G+GF+ +M+ RNTAG V HQAVA
Sbjct: 921 TYLMMIGDGINKTIITGNHSVVDGWTTFGSPTFAVVGQGFVGVNMTIRNTAGAVKHQAVA 1100
Query: 350 LRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRA 409
LR +D S FY CS EG+QDTLY HSLRQFYREC+IYGT+DFIFGN V QNC +Y R
Sbjct: 1101LRNGADLSTFYSCSFEGYQDTLYTHSLRQFYRECDIYGTVDFIFGNAKVVFQNCNLYPRL 1280
Query: 410 PLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTVYI 460
P+ Q +TAQGR P+Q TG +I + I AT TYLGRPWK+YSRTVY+
Sbjct: 1281PMSGQFNAITAQGRTDPNQDTGTSIHNCTIKATDDLAASNGAVSTYLGRPWKEYSRTVYM 1460
Query: 461 NTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYF 520
T+M ++ GW W G+FAL+TL+Y E+ N GPG+S GRV W GYH++ +A A+ F
Sbjct: 1461QTFMDNVINVAGWRAWDGEFALSTLYYAEFNNSGPGSSTDGRVTWQGYHVI-NATDAANF 1637
Query: 521 TVQRFIHGDSWLPGTGVKFTAGL 543
TV F+ GD WLP TGV +T L
Sbjct: 1638TVANFLLGDDWLPQTGVSYTNSL 1706
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 301 bits (770), Expect = 6e-82
Identities = 178/551 (32%), Positives = 285/551 (51%), Gaps = 15/551 (2%)
Frame = +1
Query: 7 LFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNELTKIGP 66
L + M+ ++ S +K E + I Q + C + + +C H L K
Sbjct: 73 LLVAMVGVVAVSLTKGGDGEQKAHISNS--QKNVDMLCQSTKFKETC----HKTLEKASF 234
Query: 67 PSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLG 126
+ + + AL T E + I+N + + + + A+ C E++D++V + S+G
Sbjct: 235 SNMKNRIKGALGATEEELRKHINNSALYQELATDSMTKQAMEICNEVLDYAVDGIHKSVG 414
Query: 127 EMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQVTQLISN 186
+ + S +Y +++ WL+ LS+Q TCL+GF T ++ L +L SN
Sbjct: 415 TLDQFDFHKLS-EYAFDIKVWLTGTLSHQQTCLDGFVNTKTHAGETMAKVLKTSMELSSN 591
Query: 187 VLSLYTQLHAL--PFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVV 244
+ + + + F P + + S L D P W+++G + LL +A+AVV
Sbjct: 592 AIDMMDVVSRILKGFHPSQ---YGVSRRLLSDDGIPSWVSDGHRHLLAGG--NVKANAVV 756
Query: 245 ALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTII 304
A DGSG ++T+T+A+ P + +VIYVK G+Y+E V++ ++M + ++GDG +T
Sbjct: 757 AQDGSGQFKTLTDALKTVPPTNAAPFVIYVKAGVYKETVNVAKEMNYVTVIGDGPTKTKF 936
Query: 305 TSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSI 364
T + N+ G T++TAT V+G F+A+D+ F NTAG QAVALRV +DQ+ F+ C +
Sbjct: 937 TGSLNYADGINTYKTATFGVNGANFMAKDIGFENTAGTSKFQAVALRVTADQAIFHNCQM 1116
Query: 365 EGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRK 424
+GFQDTL+ S RQFYR+C I GTIDF+FG+ V QNCK+ R P QK VTA GR
Sbjct: 1117DGFQDTLFVESQRQFYRDCAISGTIDFVFGDAFGVFQNCKLICRVPAKGQKCLVTAGGRD 1296
Query: 425 SPHQSTGFTIQDSFI--------LATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEW 476
+ ++ S + + +YLGRPWK YS+ V +++ + M P G++
Sbjct: 1297KQNSASALVFLSSHFTGEPALTSVTPKLSYLGRPWKLYSKVVIMDSTIDAMFAPEGYMPM 1476
Query: 477 FGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASY-----FTVQRFIHGDSW 531
G T + EY N GPGA RVKW G ++ A Y F + D+W
Sbjct: 1477VGGAFKDTCTFYEYNNKGPGADTNLRVKWHGVKVLTSNVAAEYYPGKFFEIVNATARDTW 1656
Query: 532 LPGTGVKFTAG 542
+ +GV ++ G
Sbjct: 1657IVKSGVPYSLG 1689
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 249 bits (636), Expect = 2e-66
Identities = 119/220 (54%), Positives = 160/220 (72%), Gaps = 1/220 (0%)
Frame = +3
Query: 212 SLDLDSEFPEWMTEGDQELLRSKPH-RSRADAVVALDGSGHYRTITEAVNAAPSHSNRRY 270
SL +E P W+ D++LL+ + +AD VVA DGSG Y+TI+ A+ P+ S++R
Sbjct: 30 SLSHQNEEPNWLHHKDRKLLQKDSDLKKKADIVVAKDGSGKYKTISAALKHVPNKSDKRT 209
Query: 271 VIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFI 330
VIYVKKG+Y ENV +++ N+M++GDG+ TI++ + NF+ G TF TAT AV G+ FI
Sbjct: 210 VIYVKKGIYYENVRVEKTKWNVMIIGDGMNVTIVSGSLNFIDGTPTFSTATFAVFGRNFI 389
Query: 331 ARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTID 390
ARD+ F+NTAGP HQAVAL +DQ+ FY+CS++ +QDTLYAHS RQFYREC IYGT+D
Sbjct: 390 ARDIGFKNTAGPQKHQAVALMTSADQAVFYKCSMDAYQDTLYAHSNRQFYRECNIYGTVD 569
Query: 391 FIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQST 430
FIFGN A VLQNC I R P+P Q+ T+TAQG+ P+ +T
Sbjct: 570 FIFGNSAVVLQNCNILPRQPMPGQQNTITAQGKTDPNMNT 689
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 217 bits (552), Expect(2) = 4e-61
Identities = 102/161 (63%), Positives = 130/161 (80%)
Frame = +2
Query: 248 GSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSN 307
GSG Y + +AV+AAP S +RYVIYVKKG+Y ENV++K+K NIML+G+G+ TII+ +
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGS 181
Query: 308 RNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGF 367
RN++ G TTFR+AT AVSG+GFIARD+SF+NTAG HQAVALR DSD S FYRC I G+
Sbjct: 182 RNYVDGSTTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCGIFGY 361
Query: 368 QDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTR 408
QD+LY H++RQFYREC+I GT+DFIFG+ AV QNC+I +
Sbjct: 362 QDSLYTHTMRQFYRECKISGTVDFIFGDATAVFQNCQILAK 484
Score = 36.6 bits (83), Expect(2) = 4e-61
Identities = 16/23 (69%), Positives = 19/23 (82%)
Frame = +3
Query: 411 LPLQKVTVTAQGRKSPHQSTGFT 433
+P QK TVTAQGRK P+Q TGF+
Sbjct: 492 MPKQKNTVTAQGRKDPNQPTGFS 560
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 204 bits (519), Expect = 7e-53
Identities = 108/218 (49%), Positives = 138/218 (62%), Gaps = 20/218 (9%)
Frame = +1
Query: 257 EAVNAAPSHS--NRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQ-G 313
EAVNAAP + +R+VIY+K+G+Y E V + K N++ +GDGIG+T+IT + N Q G
Sbjct: 4 EAVNAAPDNGVDRKRFVIYIKEGVYEETVRVPLKKRNVVFLGDGIGKTVITGSANVGQPG 183
Query: 314 WTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYA 373
TT+ +ATVAV G GF+A+D++ NTAGP HQAVA R+DSD S C G QDTLYA
Sbjct: 184 MTTYNSATVAVLGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLYA 363
Query: 374 HSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTR----APLPLQKVTVTAQGRKSPHQS 429
HSLRQFY+ C I G +DFIFGN AA+ Q+C+I R P + +TA GR P QS
Sbjct: 364 HSLRQFYKSCRIVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTDPAQS 543
Query: 430 TGFTIQDSFILATQ-------------PTYLGRPWKQY 454
TGF Q+ I T+ YLGRPWK+Y
Sbjct: 544 TGFVFQNCLINGTEDYMALYHSNPKVHKNYLGRPWKEY 657
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 143 bits (361), Expect(2) = 2e-46
Identities = 83/237 (35%), Positives = 134/237 (56%), Gaps = 12/237 (5%)
Frame = +3
Query: 227 DQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMK 286
D +L +++ ++ R V+ DGS +++ITEA+N+ ++ RR +I + G YRE + +
Sbjct: 198 DPKLKKAESNKVRLK--VSQDGSAQFKSITEALNSIQPYNIRRVIISIAPGYYREKIVVP 371
Query: 287 RKMTNIMLVGDGIGQTIITSNRNFMQGWT--------TFRTATVAVSGKGFIARDMSFRN 338
+ + I +GD IT N Q T TF +ATVAV+ F+A +++F N
Sbjct: 372 KTLPFITFLGDVRDPPTITGNDT--QSVTGSDGAQLRTFNSATVAVNASYFMAININFEN 545
Query: 339 TA----GPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFG 394
TA G QAVA+R+ +++AFY C+ G QDTLY H ++ C I G++DFI G
Sbjct: 546 TASFPIGSKVEQAVAVRITGNKTAFYNCTFSGVQDTLYDHKGLHYFNNCTIKGSVDFICG 725
Query: 395 NGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQPTYLGRPW 451
+G ++ + C I + A ++TAQ +P +GF+ ++S ++ PTYLGRPW
Sbjct: 726 HGKSLYEGCTIRSIAN---NMTSITAQSGSNPSYDSGFSFKNSMVIGDGPTYLGRPW 887
Score = 61.2 bits (147), Expect(2) = 2e-46
Identities = 30/79 (37%), Positives = 43/79 (53%)
Frame = +1
Query: 454 YSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKD 513
YS+ V+ TYM V P+GW +W +YGEY+ GPG++ AGRV W M +
Sbjct: 895 YSQVVFSYTYMDNSVLPKGWEDWNDTKRYMNAYYGEYKCSGPGSNTAGRVPWAR---MLN 1065
Query: 514 AATASYFTVQRFIHGDSWL 532
A F ++I G++WL
Sbjct: 1066DKEAQVFIGTQYIDGNTWL 1122
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 174 bits (440), Expect = 1e-43
Identities = 96/218 (44%), Positives = 124/218 (56%), Gaps = 14/218 (6%)
Frame = +1
Query: 269 RYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKG 328
RY IYVK G+Y E + + + NI++ GDG G+TI+T +N G T +TAT A + G
Sbjct: 4 RYTIYVKAGVYDEYITIPKDAVNILMYGDGPGKTIVTGRKNGAAGVKTMQTATFANTALG 183
Query: 329 FIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGT 388
FI + M+F NTAGP HQAVA R D SA C I G+QDTLY + RQFYR C I GT
Sbjct: 184 FIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVISGT 363
Query: 389 IDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILA------- 441
+DFIFG A ++Q+ I R P P Q T+TA G +TG IQ I+
Sbjct: 364 VDFIFGTSATLIQDSTIIVRMPSPNQFNTITADGSYVNKLNTGIVIQGCNIVPKAALFPQ 543
Query: 442 --TQPTYLGRPWKQYSRTVYINT-----YMSGMVQPRG 472
T +YLGRPWK ++TV + + Y S + P G
Sbjct: 544 RFTIKSYLGRPWKVLTKTVVMESTIG*FYSSXWMDPLG 657
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 173 bits (439), Expect = 1e-43
Identities = 84/200 (42%), Positives = 116/200 (58%), Gaps = 8/200 (4%)
Frame = +2
Query: 329 FIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGT 388
F A ++ F N+AG HQAVALRV +D++ FY C + G+QDTLY S RQFYR+C I GT
Sbjct: 86 FTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCTITGT 265
Query: 389 IDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSF------ILAT 442
IDF+FG+ V QNCK+ R P+ Q+ VTA GR + Q+ +L
Sbjct: 266 IDFVFGDAVGVFQNCKLIVRKPMANQQCMVTAGGRTKVDSVSALVFQNCHFTGEPEVLTM 445
Query: 443 QP--TYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLA 500
QP YLGRPW+ +S+ V +++ + G+ P G++ W G+ T Y EY N G GA+
Sbjct: 446 QPKIAYLGRPWRNFSKVVIVDSLIDGLFVPEGYMPWMGNLFKETCTYLEYNNKGAGAATN 625
Query: 501 GRVKWPGYHIMKDAATASYF 520
+VKWPG + A Y+
Sbjct: 626 LKVKWPGVKTISAGEAAKYY 685
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 165 bits (418), Expect = 4e-41
Identities = 78/144 (54%), Positives = 108/144 (74%), Gaps = 3/144 (2%)
Frame = +1
Query: 247 DGSGHYRTITEAVNAAPSH---SNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTI 303
DGSG++ TI +A+ AAP++ S+ ++I++ +G+Y E V + +K +M++G+GI QTI
Sbjct: 64 DGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEYVSIDKKKKYLMMIGEGINQTI 243
Query: 304 ITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCS 363
IT NRN G+TTF +AT AV +GF+A +++FRNTAG +QAVALR +D S FY CS
Sbjct: 244 ITGNRNVADGFTTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFYSCS 423
Query: 364 IEGFQDTLYAHSLRQFYRECEIYG 387
EG+QDTLY HSLRQFYREC+IYG
Sbjct: 424 FEGYQDTLYTHSLRQFYRECDIYG 495
>TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE8B
precursor (EC 3.1.1.11) (Pectin methylesterase) (PE).
[Peach] {Prunus persica}, partial (34%)
Length = 1415
Score = 158 bits (400), Expect = 4e-39
Identities = 102/319 (31%), Positives = 166/319 (51%), Gaps = 1/319 (0%)
Frame = +1
Query: 4 VNALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNELTK 63
++ ++L++ +I SS T + I + + +S + N+
Sbjct: 19 ISDTYLLIVSIIFQSSKMELCSSKQPSTTTLSITTFIFFFLLLLSLPHSTFASSPNDFVG 198
Query: 64 IGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAW 123
T+ + + + Q I L++F+ +R A+ DC +L+D S+ +L
Sbjct: 199 SSLRVSTAKFANSAEEVITVLQKVISILSRFTNVFGHSRTSNAVSDCLDLLDMSLDQLNQ 378
Query: 124 SLGEMRRIRAGDTSV-QYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQVTQ 182
S+ ++ + D S + +L WLSA L DTC+EG EG+ ++ IS L V
Sbjct: 379 SISAAQKPKEKDNSTGKLNCDLRTWLSAVLVYPDTCIEGLEGSI--VKGLISSGLDHVMS 552
Query: 183 LISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADA 242
L++N+L N + + + D FP W+ + D +LL++ + ADA
Sbjct: 553 LVANLLGEVVS----------GNDDQLATNKD---RFPSWIRDEDTKLLQA--NGVTADA 687
Query: 243 VVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQT 302
VVA DGSG Y + +AV+AAP S +RYVIYVKKG+Y ENV++K+K NIML+G+G+ T
Sbjct: 688 VVAADGSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDAT 867
Query: 303 IITSNRNFMQGWTTFRTAT 321
II+ +RN++ G TTFR+AT
Sbjct: 868 IISGSRNYVDGSTTFRSAT 924
>TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein -
Arabidopsis thaliana, partial (18%)
Length = 467
Score = 137 bits (346), Expect = 8e-33
Identities = 58/98 (59%), Positives = 76/98 (77%)
Frame = +2
Query: 446 YLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKW 505
YLGRPWK YSRT+++ +YMS ++P GWLEW G+FAL TL+Y EY N GPGA +A RVKW
Sbjct: 2 YLGRPWKTYSRTIFMQSYMSDAIRPEGWLEWNGNFALNTLYYAEYMNSGPGAGVANRVKW 181
Query: 506 PGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
GYH++ D++ A+ FTV +FI G+ LP TGV +T+GL
Sbjct: 182 SGYHVLNDSSEATKFTVAQFIEGNLGLPSTGVTYTSGL 295
>TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 483
Score = 135 bits (340), Expect = 4e-32
Identities = 59/104 (56%), Positives = 75/104 (71%)
Frame = +1
Query: 440 LATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASL 499
+AT TYLGRPWK+YSRTVY+ ++M + P GW EW GDFAL+TL+Y EY N G G+S
Sbjct: 4 VATVKTYLGRPWKEYSRTVYMQSFMDSFINPSGWREWNGDFALSTLYYAEYDNRGAGSST 183
Query: 500 AGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
A RV WPGYH++ A A+ FTV F+ GD W+P TGV + +GL
Sbjct: 184 ANRVTWPGYHVI-GATDAANFTVSNFVSGDDWIPQTGVPYMSGL 312
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,915,864
Number of Sequences: 36976
Number of extensions: 212052
Number of successful extensions: 1062
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 1002
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1005
length of query: 545
length of database: 9,014,727
effective HSP length: 101
effective length of query: 444
effective length of database: 5,280,151
effective search space: 2344387044
effective search space used: 2344387044
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0109.6


