

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0109.3
(297 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
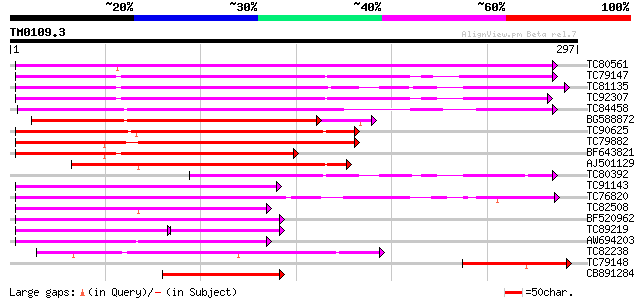
Score E
Sequences producing significant alignments: (bits) Value
TC80561 weakly similar to GP|10177856|dbj|BAB11208. phloem-speci... 218 3e-57
TC79147 weakly similar to GP|10177856|dbj|BAB11208. phloem-speci... 212 2e-55
TC81135 weakly similar to GP|10177856|dbj|BAB11208. phloem-speci... 204 3e-53
TC92307 weakly similar to PIR|E84434|E84434 probable phloem-spec... 202 1e-52
TC84458 weakly similar to PIR|B96604|B96604 hypothetical protein... 201 4e-52
BG588872 weakly similar to GP|21592660|gb phloem-specific lectin... 167 3e-45
TC90625 weakly similar to GP|10177856|dbj|BAB11208. phloem-speci... 162 2e-40
TC79882 weakly similar to PIR|E84434|E84434 probable phloem-spec... 160 7e-40
BF643821 weakly similar to PIR|F84434|F844 lectin-like protein [... 140 6e-34
AJ501129 weakly similar to GP|10177856|dbj phloem-specific lecti... 126 9e-30
TC80392 weakly similar to GP|15088626|gb|AAK84134.1 nictaba {Nic... 126 9e-30
TC91143 similar to PIR|T47557|T47557 hypothetical protein F8J2.1... 107 6e-24
TC76820 weakly similar to GP|18175688|gb|AAL59911.1 unknown prot... 105 2e-23
TC82508 similar to PIR|T50527|T50527 hypothetical protein T27I15... 93 1e-19
BF520962 weakly similar to PIR|C96656|C96 unknown protein 43836... 92 3e-19
TC89219 weakly similar to PIR|C96656|C96656 unknown protein 438... 66 1e-18
AW694203 weakly similar to PIR|C96656|C96 unknown protein 43836... 79 2e-15
TC82238 weakly similar to GP|8570046|dbj|BAA96751.1 Similar to A... 72 3e-13
TC79148 57 9e-09
CB891284 weakly similar to GP|22122920|gb unknown protein {Oryza... 54 7e-08
>TC80561 weakly similar to GP|10177856|dbj|BAB11208. phloem-specific
lectin-like protein {Arabidopsis thaliana}, partial
(52%)
Length = 1436
Score = 218 bits (554), Expect = 3e-57
Identities = 127/291 (43%), Positives = 171/291 (58%), Gaps = 7/291 (2%)
Frame = +3
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSR--AVNPSA 61
LPE C+ +LS T+P D R SLVSST RSAA+SD VW FLPSDY I+S+ A ++
Sbjct: 171 LPEGCIENVLSFTTPRDVARLSLVSSTFRSAAESDSVWDKFLPSDYHSIISKSDAEIDNS 350
Query: 62 LQFSSYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRS 120
L K L+ L PLL++ G KSF+LDK+ GKK Y+LSAR L I W P +W W
Sbjct: 351 LSVLPKKDLYVNLTQKPLLIENGKKSFQLDKVDGKKCYMLSARSLFIVWGDTPRYWRWIP 530
Query: 121 NPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRG-YGLDSAPAEVSV 179
+P+SRF E AEL +V W EI G I T +L+P T YGAYL+ K S G +G + P E S+
Sbjct: 531 DPDSRFPEAAELVSVCWFEIRGWISTIMLSPKTLYGAYLVFKSSAAGAFGFEYQPCEASI 710
Query: 180 AVGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLE-QLEEEGKDGGVIPVP 238
+ G +L +++ + + I +E +E + P
Sbjct: 711 DIAGGDTVERNVFLDAGRGRRLRYQIVPRSRTTGILTRLRSPVEAPVEPTESVADLRKYP 890
Query: 239 GKRDDGWMEIELGEFFS-GGGDVEIKMGLREV-GYQLKGGLVVEGIEVRPK 287
+R DGW+E+ELGEFF+ GG D ++ +G+ EV G KGGLVV+GIE+RPK
Sbjct: 891 KERADGWLEMELGEFFNEGGDDKQVDIGVCEVKGGGWKGGLVVQGIEIRPK 1043
>TC79147 weakly similar to GP|10177856|dbj|BAB11208. phloem-specific
lectin-like protein {Arabidopsis thaliana}, partial
(32%)
Length = 958
Score = 212 bits (539), Expect = 2e-55
Identities = 126/287 (43%), Positives = 165/287 (56%), Gaps = 3/287 (1%)
Frame = +2
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
L E C+S ILS T+P DA R S+VS T SAADSD VW FLPSD I+S++ PS
Sbjct: 41 LAEGCISSILSRTTPVDAGRLSVVSKTFLSAADSDAVWNHFLPSDVNSIISQS--PSLAN 214
Query: 64 FSSYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNP 122
S K L+ L P+++D KSF+LD+ SGKK Y+L+AR L I W D +W W + P
Sbjct: 215 APSKKALYLTLSDRPIIIDDAKKSFQLDRKSGKKCYMLAARSLHIIWGDDDRYWIWTAMP 394
Query: 123 ESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAV- 181
+SRF EVA LR V WLEI G I L+PNT Y AYL+ K+ GYG ++ P ++SV V
Sbjct: 395 DSRFPEVANLRLVWWLEIRGMINNLALSPNTQYAAYLVFKLID-GYGFETLPVDLSVGVE 571
Query: 182 GGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKR 241
GG + + ++ FYG +RV+ L P PG R
Sbjct: 572 GGHSSTKIVCLDPNVERRQNSRHARFYGR-----ADRVVGL-------------PRPGVR 697
Query: 242 DDGWMEIELGEFFSGGGDVEIKMGLREV-GYQLKGGLVVEGIEVRPK 287
DGW+EIE+GEF SG + E++M + E+ + KG +EGIEVRPK
Sbjct: 698 SDGWLEIEMGEFNSGLENEEVQMSVIEIKAGETKGNFFLEGIEVRPK 838
>TC81135 weakly similar to GP|10177856|dbj|BAB11208. phloem-specific
lectin-like protein {Arabidopsis thaliana}, partial
(32%)
Length = 1186
Score = 204 bits (520), Expect = 3e-53
Identities = 126/293 (43%), Positives = 170/293 (58%), Gaps = 3/293 (1%)
Frame = +2
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE C++ I+S T+P DA R S+ S T RSA+DSD VW FLPSD I+S + PS
Sbjct: 116 LPEGCIAAIVSRTTPLDAGRLSVFSKTFRSASDSDAVWNQFLPSDISSIISHS--PSLTN 289
Query: 64 FSSYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNP 122
+ K L+ AL P+++D KS +L++ SGKK Y+LSAR L+I W D +W+W S P
Sbjct: 290 IPTKKDLYLALSDRPVIIDHDKKSIQLERKSGKKCYMLSARSLAIVWGDDRRYWNWISMP 469
Query: 123 ESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVG 182
+SRF EVA+L V WLEI G I T VL+PNT Y AY++ K+ + G + PA++SV V
Sbjct: 470 DSRFPEVAKLVDVCWLEIHGVINTIVLSPNTQYAAYVVFKMINAS-GFQNRPADLSVGVE 646
Query: 183 GRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKRD 242
G GQ K V ++ G R +RV L++ P R
Sbjct: 647 G----------GQSSTKIVCLDPNVEG--RPQLHDRVDGLQR-------------PSVRS 751
Query: 243 DGWMEIELGEFFSGG-GDVEIKMG-LREVGYQLKGGLVVEGIEVRPKQTQSTQ 293
DGW+EIE+GEFF+ G + E++M ++ G K GLV+EGIEVRPK Q
Sbjct: 752 DGWLEIEMGEFFNSGIENEEVQMNVIQTKGGNWKRGLVLEGIEVRPKYNTINQ 910
>TC92307 weakly similar to PIR|E84434|E84434 probable phloem-specific lectin
[imported] - Arabidopsis thaliana, partial (19%)
Length = 817
Score = 202 bits (515), Expect = 1e-52
Identities = 120/284 (42%), Positives = 162/284 (56%), Gaps = 3/284 (1%)
Frame = +1
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE C++ +LS T+P DACR S +S SA+DSD VW FLPSD I+S+ PS
Sbjct: 28 LPEGCIAAVLSSTTPVDACRLSFISKNFLSASDSDAVWNRFLPSDISSIISQF--PSLSN 201
Query: 64 FSSYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNP 122
+ K L+ AL P+++D G KSF+L++ SGKK Y+LSAR L+ITW +W+W P
Sbjct: 202 IPTKKDLYIALSDRPIIIDNGKKSFQLERKSGKKCYMLSARSLAITWGDTDQYWNWTVMP 381
Query: 123 ESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVG 182
ESRF EVAEL V WLEI GK T L+PNT Y YL+ K+ + YG + P E+SV +
Sbjct: 382 ESRFPEVAELLHVCWLEIRGK*NTLALSPNTRYVTYLVFKMIN-AYGFEYFPVELSVGIE 558
Query: 183 GRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKRD 242
G L + ++ + NRV++L++ P R
Sbjct: 559 GGHSSTKIVCLADPNARRRHRSRIIVTE-----PNRVLRLQR-------------PNVRS 684
Query: 243 DGWMEIELGEFF-SGGGDVEIKMGLREV-GYQLKGGLVVEGIEV 284
DGW+EIE+GEFF SG D E++M + ++ G K G VEGIEV
Sbjct: 685 DGWLEIEMGEFFISGLEDEEVQMSVIDIKGGHWKRGFFVEGIEV 816
>TC84458 weakly similar to PIR|B96604|B96604 hypothetical protein F14G9.14
[imported] - Arabidopsis thaliana, partial (24%)
Length = 766
Score = 201 bits (510), Expect = 4e-52
Identities = 123/285 (43%), Positives = 156/285 (54%), Gaps = 2/285 (0%)
Frame = +1
Query: 5 PEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQF 64
PEDCV+ ILS TSP D CR SL+SS ++S ADSD VW FLP +Y+ IVSR V+P L
Sbjct: 28 PEDCVAHILSFTSPIDVCRVSLISSIVKSMADSDNVWEKFLPHNYQGIVSRLVDPP-LSC 204
Query: 65 SSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNPES 124
SS K+L LC P L+D GNK F ++K +GK Y LSAR LSIT+ ++P+ WSWR S
Sbjct: 205 SSKKELLARLCKPQLIDDGNKMFSIEKTTGKICYSLSARQLSITFGNNPLHWSWRQVQGS 384
Query: 125 RFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVGGR 184
RF EVAELRT+ +LE G I +L+P T YGAYL +K YGLD P
Sbjct: 385 RFAEVAELRTICYLETIGSINNEMLSPKTMYGAYLKVKNVECAYGLDLLPC--------- 537
Query: 185 VLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKRDDG 244
R + RN + E +E
Sbjct: 538 -------------------------IRSNCKRNGICNCEHKDE----------------- 591
Query: 245 WMEIELGEFFSGGGDV-EIKMGLREV-GYQLKGGLVVEGIEVRPK 287
W+EIELG F++ V E++M L+EV G KG L+V+GIE+RPK
Sbjct: 592 WLEIELGSFYTEKVQVQEVRMCLKEVNGVDFKGRLIVDGIELRPK 726
>BG588872 weakly similar to GP|21592660|gb phloem-specific lectin PP2-like
protein {Arabidopsis thaliana}, partial (55%)
Length = 655
Score = 167 bits (422), Expect(2) = 3e-45
Identities = 82/152 (53%), Positives = 108/152 (70%)
Frame = +1
Query: 12 ILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQFSSYKQLF 71
ILS TSP D CR SL+SS ++S ADSD +W FLP Y++I+SR V+P L SS K+LF
Sbjct: 19 ILSFTSPLDVCRVSLLSSVVQSMADSDFLWEKFLPHKYQEIISRLVDPP-LPCSSKKELF 195
Query: 72 HALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNPESRFREVAE 131
LC P L+D GNK F ++K +GK Y+LSAR LSIT+ + ++WSW+ SRF E AE
Sbjct: 196 ARLCKPSLIDDGNKMFSIEKTTGKICYLLSARQLSITFGNTSLYWSWKQVQGSRFAEAAE 375
Query: 132 LRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKV 163
LRT+ WLEI+G I + +L+P T YGAYL +K+
Sbjct: 376 LRTICWLEIKGSINSEMLSPKTMYGAYLKVKI 471
Score = 32.3 bits (72), Expect(2) = 3e-45
Identities = 18/42 (42%), Positives = 23/42 (53%), Gaps = 11/42 (26%)
Frame = +2
Query: 162 KVSHRGYGLDSAPAEVSVAVG-----------GRVLRRGKAY 192
K++ R YGLDS P+EVS+ VG + R GKAY
Sbjct: 467 KLADRAYGLDSLPSEVSIEVGNYKSQENVYIRSQSKRNGKAY 592
>TC90625 weakly similar to GP|10177856|dbj|BAB11208. phloem-specific
lectin-like protein {Arabidopsis thaliana}, partial (9%)
Length = 892
Score = 162 bits (409), Expect = 2e-40
Identities = 91/184 (49%), Positives = 117/184 (63%), Gaps = 4/184 (2%)
Frame = +2
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE C+++ILSHT+P D+CR SLV SAA SD VW FLPSD I+S + + S+L
Sbjct: 233 LPEGCIAKILSHTAPVDSCRLSLVCKGFCSAAKSDTVWDRFLPSDLISIISDSPSASSL- 409
Query: 64 FS---SYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWR 119
FS S K L+ L P++++ G KSF+L+K SG+K Y+LSARD+SI P FW W
Sbjct: 410 FSTSPSKKSLYLTLSDHPIVIENGKKSFQLEKQSGRKIYMLSARDISIALGDTPQFWDWP 589
Query: 120 SNPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSV 179
PESRFREVA LR V W EG I VL+ NT Y A+L+ + S +G D P E++V
Sbjct: 590 ILPESRFREVARLRIVCWFAFEGTINKHVLSSNTQYAAFLVSR*S-MPFGFDEIPIELTV 766
Query: 180 AVGG 183
V G
Sbjct: 767 GVLG 778
>TC79882 weakly similar to PIR|E84434|E84434 probable phloem-specific lectin
[imported] - Arabidopsis thaliana, partial (14%)
Length = 636
Score = 160 bits (404), Expect = 7e-40
Identities = 89/185 (48%), Positives = 116/185 (62%), Gaps = 5/185 (2%)
Frame = +1
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSD---YEDIVSRAVNPS 60
LPE C+S ILS T+P DA R S+VS T+ SAADSD VW FLPSD + I+S A P+
Sbjct: 79 LPEGCISAILSRTTPADAGRLSVVSKTILSAADSDAVWNQFLPSDSHFIDSIISLANVPT 258
Query: 61 ALQFSSYKQLFHALC-SPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWR 119
K L+ AL SP+++D KS +LD+ SGKK Y+L+AR LSI W +D +W+W
Sbjct: 259 K------KSLYLALSDSPIIIDNDQKSIQLDRKSGKKCYMLAARSLSIAWGNDDRYWNWI 420
Query: 120 SNPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMK-VSHRGYGLDSAPAEVS 178
+ P+SRF EVAEL V WL I G I L+PNT Y AYL+ K + G+ + P +S
Sbjct: 421 AMPDSRFPEVAELLVVCWLHISGMINMLALSPNTQYAAYLVFKMIGGFGFRNPNCPVVLS 600
Query: 179 VAVGG 183
+ V G
Sbjct: 601 ICVEG 615
>BF643821 weakly similar to PIR|F84434|F844 lectin-like protein [imported] -
Arabidopsis thaliana, partial (14%)
Length = 584
Score = 140 bits (353), Expect = 6e-34
Identities = 78/152 (51%), Positives = 96/152 (62%), Gaps = 4/152 (2%)
Frame = +2
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSD---YEDIVSRAVNPS 60
L E C+S I+S T+P DA R S+VS T SAADSD VW FLPSD + I+S + PS
Sbjct: 92 LSEGCISAIISRTTPADAGRLSVVSKTFLSAADSDAVWNQFLPSDSHFIDSIISHS--PS 265
Query: 61 ALQFSSYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWR 119
S K L+ AL P+++D KSF+LD+ SGKK Y+L+AR LSI W D +W+W
Sbjct: 266 LANVPSKKALYLALSDLPIIIDNDQKSFQLDRKSGKKCYMLAARSLSIAWGDDRRYWNWI 445
Query: 120 SNPESRFREVAELRTVNWLEIEGKIRTGVLTP 151
S P SRF EVAEL V WL+I G I T L P
Sbjct: 446 SMPNSRFPEVAELLEVWWLDIRGMINTLGLVP 541
>AJ501129 weakly similar to GP|10177856|dbj phloem-specific lectin-like
protein {Arabidopsis thaliana}, partial (33%)
Length = 558
Score = 126 bits (317), Expect = 9e-30
Identities = 70/150 (46%), Positives = 96/150 (63%), Gaps = 3/150 (2%)
Frame = +2
Query: 33 SAADSDMVWRSFLPSDYEDIVSRAVNPSALQFSS--YKQLFHALCS-PLLLDGGNKSFKL 89
SAA+SD+VW FLPSD IVS + + S+L +S K L+ L P+L++ G SF+L
Sbjct: 107 SAAESDIVWDRFLPSDLISIVSDSQSASSLFTTSPSKKSLYLTLSDHPILINNGKTSFQL 286
Query: 90 DKLSGKKSYILSARDLSITWSSDPMFWSWRSNPESRFREVAELRTVNWLEIEGKIRTGVL 149
+K SGKK Y+L ARDLSI W P +W W PESRF+EVA LRTV W EI G I VL
Sbjct: 287 EKQSGKKVYMLGARDLSIAWGDTPCYWDWIILPESRFQEVARLRTVCWFEISGTINKRVL 466
Query: 150 TPNTSYGAYLIMKVSHRGYGLDSAPAEVSV 179
+ ++ Y A+L+ K+ + Y + P +++V
Sbjct: 467 SSDSQYVAFLVFKMIN-AYRFEDLPTKLTV 553
>TC80392 weakly similar to GP|15088626|gb|AAK84134.1 nictaba {Nicotiana
tabacum}, partial (29%)
Length = 1161
Score = 126 bits (317), Expect = 9e-30
Identities = 79/193 (40%), Positives = 104/193 (52%)
Frame = +2
Query: 95 KKSYILSARDLSITWSSDPMFWSWRSNPESRFREVAELRTVNWLEIEGKIRTGVLTPNTS 154
K+S +LSAR LSI W D +W+W + P+SRF EVA+L V WL+I G I T L+PNT
Sbjct: 83 KESCMLSARSLSIAWGDDKRYWNWINMPDSRFPEVAQLLKVWWLDISGTINTLSLSPNTQ 262
Query: 155 YGAYLIMKVSHRGYGLDSAPAEVSVAVGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDI 214
Y AYL+ K+ G + P E+SV V G GQ K ++ G +
Sbjct: 263 YAAYLVFKMI-EAEGFQNCPIEISVGVEG----------GQSGTKIDCLDPNVEGGQ--- 400
Query: 215 FRNRVIQLEQLEEEGKDGGVIPVPGKRDDGWMEIELGEFFSGGGDVEIKMGLREVGYQLK 274
NR + L++ P R DGW+EIE+GEF SG D ++ M ++E K
Sbjct: 401 -HNRAVGLQR-------------PSVRSDGWLEIEMGEFNSGLEDGDVLMNVKETN-NWK 535
Query: 275 GGLVVEGIEVRPK 287
GL VEGIEVRPK
Sbjct: 536 SGLFVEGIEVRPK 574
>TC91143 similar to PIR|T47557|T47557 hypothetical protein F8J2.170 -
Arabidopsis thaliana, partial (60%)
Length = 669
Score = 107 bits (267), Expect = 6e-24
Identities = 50/139 (35%), Positives = 78/139 (55%)
Frame = +1
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
+PE+CV+ + H +PP+ C + ++ R AA +D VW S LPS+Y+ ++ +
Sbjct: 157 IPENCVARVFLHLTPPEICNLARLNRAFRGAASADSVWESKLPSNYQHLLRLMPEEQRCR 336
Query: 64 FSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNPE 123
S K +F PL D GNK LD+++G+ +SA+ LSIT D +W+W E
Sbjct: 337 NLSKKDIFAVFSKPLPFDDGNKQLWLDRVTGRVCMSISAKGLSITGIDDRRYWTWVPTEE 516
Query: 124 SRFREVAELRTVNWLEIEG 142
SRF VA L+ + W E++G
Sbjct: 517 SRFNIVAYLQQIWWFEVDG 573
>TC76820 weakly similar to GP|18175688|gb|AAL59911.1 unknown protein
{Arabidopsis thaliana}, partial (67%)
Length = 1388
Score = 105 bits (263), Expect = 2e-23
Identities = 73/288 (25%), Positives = 128/288 (44%), Gaps = 3/288 (1%)
Frame = +2
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
+PE C+S I+ P + C+ + V+ T A+ D VW S LP Y+ +V + + L
Sbjct: 197 IPESCISSIMMRLDPQEICKLARVNKTFHRASSDDYVWESKLPPSYKFLVKKILGEEKLC 376
Query: 64 FSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNPE 123
+ K+++ LC P DGG K LD+ SG+ +S++ IT D +W++ S E
Sbjct: 377 SMTKKEIYAKLCQPNFFDGGTKEILLDRCSGQVCLFISSKSFKITGIDDRRYWNYISTEE 556
Query: 124 SRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVGG 183
SRF+ VA L+ + W+E+ G++ + P +Y + +++ + + +G
Sbjct: 557 SRFKSVAYLQQMWWVEVLGELE--LEFPKGNYSIFFKLQL-----------GKTTKRLGR 697
Query: 184 RVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKRDD 243
RV + + + ++ T N L Q G PG+
Sbjct: 698 RVCNLEQVHGWDLKPVRFQLSTSDGQN----------SLSQCYMNG--------PGE--- 814
Query: 244 GWMEIELGEFF--SGGGDVEIKMGLREVG-YQLKGGLVVEGIEVRPKQ 288
W +G+F G + IK L ++ KGGL ++G + PK+
Sbjct: 815 -WAYYHVGDFVIEKPNGLISIKFSLAQIDCTHTKGGLCIDGAVICPKE 955
>TC82508 similar to PIR|T50527|T50527 hypothetical protein T27I15_150 -
Arabidopsis thaliana, partial (71%)
Length = 1761
Score = 93.2 bits (230), Expect = 1e-19
Identities = 49/139 (35%), Positives = 73/139 (52%), Gaps = 5/139 (3%)
Frame = +2
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
+PE CV+ +L + PPD C+ + ++ R A+ +D VW S LP +YE I+ +A+
Sbjct: 719 IPESCVALVLMYLDPPDICKLARLNRAFRDASFADFVWESKLPLNYEFIMGKALEDDTTS 898
Query: 64 FSS-----YKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSW 118
SS + ++ LC P L D G K LDK +G +S++ L IT D +W+
Sbjct: 899 SSSGAELGKRDIYARLCKPNLFDNGTKEIWLDKRTGGVCLAISSKALRITGIDDRRYWNH 1078
Query: 119 RSNPESRFREVAELRTVNW 137
S ESRF VA L + W
Sbjct: 1079ISTEESRFHTVAYLHQIWW 1135
>BF520962 weakly similar to PIR|C96656|C96 unknown protein 43836-42510
[imported] - Arabidopsis thaliana, partial (58%)
Length = 681
Score = 91.7 bits (226), Expect = 3e-19
Identities = 47/142 (33%), Positives = 77/142 (54%), Gaps = 1/142 (0%)
Frame = +3
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LP+ C++ I+ + PP C+ + ++ A+ +D VW S LPS+Y I+ + + S+
Sbjct: 180 LPQSCIAGIMEYMDPPQICKLASLNRAFHGASLADFVWESKLPSNYHVIIQKIFDGSSFP 359
Query: 64 FSSYKQ-LFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNP 122
K+ ++ LCS D G K LD+ GK +SA+ LSIT D +W+
Sbjct: 360 SHLGKRGIYSRLCSLNTFDEGTKKVWLDRSMGKVCLSVSAKGLSITGIDDRRYWNHVPTE 539
Query: 123 ESRFREVAELRTVNWLEIEGKI 144
ESRF VA L+ + W E++G++
Sbjct: 540 ESRFSSVAYLQQIWWFEVDGEV 605
>TC89219 weakly similar to PIR|C96656|C96656 unknown protein 43836-42510
[imported] - Arabidopsis thaliana, partial (46%)
Length = 764
Score = 66.2 bits (160), Expect(2) = 1e-18
Identities = 30/82 (36%), Positives = 44/82 (53%)
Frame = +3
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
+PE C+S IL H P D C+ + V+ A+ +D VW S LPS Y+ + ++ + L
Sbjct: 282 IPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENLS 461
Query: 64 FSSYKQLFHALCSPLLLDGGNK 85
S K ++ LC P DGG K
Sbjct: 462 TMSKKDVYTKLCQPNRFDGGTK 527
Score = 43.9 bits (102), Expect(2) = 1e-18
Identities = 21/60 (35%), Positives = 34/60 (56%)
Frame = +1
Query: 85 KSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNPESRFREVAELRTVNWLEIEGKI 144
K LDK SG+ +S++ L IT D +W + ESR + VA L+ + W+E+ G++
Sbjct: 526 KEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPTEESRIKNVAYLQQMWWVEVIGEL 705
>AW694203 weakly similar to PIR|C96656|C96 unknown protein 43836-42510
[imported] - Arabidopsis thaliana, partial (52%)
Length = 621
Score = 79.3 bits (194), Expect = 2e-15
Identities = 40/135 (29%), Positives = 72/135 (52%), Gaps = 1/135 (0%)
Frame = +1
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE CV+ I+ + PP C+ + ++ R+A+ +D VW S LP +Y ++++ + +
Sbjct: 172 LPESCVASIIGYMDPPQICQLATLNRAFRAASSADFVWESKLPPNYHLLLAKIFHHFPIN 351
Query: 64 FSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSA-RDLSITWSSDPMFWSWRSNP 122
+ ++ +LC +D G K +D+ +GK +SA + LS+ D +W++
Sbjct: 352 LGK-RDIYASLCRLNTIDDGTKKVWIDRATGKLCLAISANKGLSVLGVDDRRYWNYIITE 528
Query: 123 ESRFREVAELRTVNW 137
ESRF VA L+ W
Sbjct: 529 ESRFNTVAYLQHTWW 573
>TC82238 weakly similar to GP|8570046|dbj|BAA96751.1 Similar to Arabidopsis
thaliana chromosome4 BAC clone T16H5; lectin like
protein (AL024486), partial (24%)
Length = 819
Score = 72.0 bits (175), Expect = 3e-13
Identities = 53/188 (28%), Positives = 90/188 (47%), Gaps = 6/188 (3%)
Frame = +3
Query: 15 HTSPPDACRFSLVSSTLR----SAADSDMVWRSFLPSDYEDIVSRAVNPSALQFSSYKQL 70
HT P + + S+ ++ +A ++ L +YE I+ A +P + SS +++
Sbjct: 174 HTKSPSSKKAVQNSTVVKCDETQSATKPLIKELSLSHNYEHILKDADSP--VDKSSREKM 347
Query: 71 FHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSW--RSNPESRFRE 128
L + + L K + ++K S ++L AR LSITW+ DP +W W + + E
Sbjct: 348 CDQLYAGVFLHHKTKKYWVEKNSKANCFMLYARALSITWAEDPNYWKWIQQKDVSEGTTE 527
Query: 129 VAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVGGRVLRR 188
VAEL+ V WLE+ GK T L+P Y I+ + G + P V + + G ++
Sbjct: 528 VAELKRVCWLEVHGKFDTRKLSPGILYQVSFIIMLKDPAQGWE-LPVNVRLVLPGGKKQQ 704
Query: 189 GKAYLGQK 196
K L +K
Sbjct: 705 HKENLMEK 728
>TC79148
Length = 553
Score = 57.0 bits (136), Expect = 9e-09
Identities = 29/60 (48%), Positives = 38/60 (63%), Gaps = 3/60 (5%)
Frame = +1
Query: 238 PGKRDDGWMEIELGEFFSGG-GDVEIKMGLREV--GYQLKGGLVVEGIEVRPKQTQSTQK 294
P R DGW+E E+GEFF+ G D E++M + + GY K G VEGIEVRPK+ + K
Sbjct: 130 PRMRSDGWLETEMGEFFNSGLEDEEVQMSITGIKDGYTWKRGFFVEGIEVRPKEDNNQYK 309
>CB891284 weakly similar to GP|22122920|gb unknown protein {Oryza sativa
(japonica cultivar-group)}, partial (37%)
Length = 588
Score = 53.9 bits (128), Expect = 7e-08
Identities = 24/64 (37%), Positives = 39/64 (60%)
Frame = +2
Query: 81 DGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNPESRFREVAELRTVNWLEI 140
DGG K LD+ SG+ +S++ IT D +W++ S ESRF+ VA L+ + W+E+
Sbjct: 236 DGGTKEILLDRCSGQVCLFISSKSFKITGIDDRRYWNYISTEESRFKSVAYLQQMWWVEV 415
Query: 141 EGKI 144
G++
Sbjct: 416 LGEL 427
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,938,575
Number of Sequences: 36976
Number of extensions: 121566
Number of successful extensions: 641
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 609
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 615
length of query: 297
length of database: 9,014,727
effective HSP length: 95
effective length of query: 202
effective length of database: 5,502,007
effective search space: 1111405414
effective search space used: 1111405414
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0109.3


