

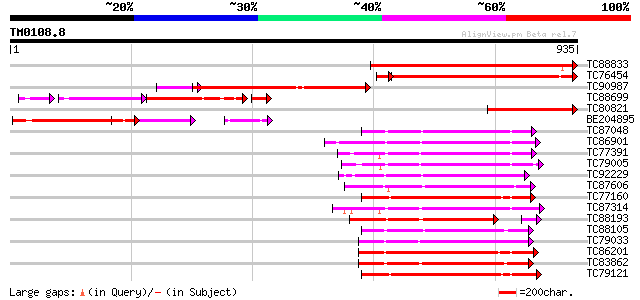
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.8
(935 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88833 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 (EC 2.7... 463 e-130
TC76454 similar to PIR|T04124|T04124 receptor-like protein kinas... 384 e-112
TC90987 similar to GP|15386483|emb|CAC60096. Seq ID: 318O13_regi... 389 e-108
TC88699 weakly similar to GP|9293948|dbj|BAB01851.1 contains sim... 158 2e-82
TC80821 similar to GP|21239384|gb|AAM44275.1 receptor-like kinas... 263 2e-70
BE204895 weakly similar to GP|15386544|em Seq ID: rhg4_Jack_ampl... 237 1e-62
TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsi... 228 1e-59
TC86901 similar to GP|13937147|gb|AAK50067.1 AT5g13160/T19L5_120... 227 1e-59
TC77391 similar to GP|14719339|gb|AAK73157.1 putative protein ki... 226 2e-59
TC79005 similar to GP|19423982|gb|AAL87287.1 putative protein ki... 226 4e-59
TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC980... 224 2e-58
TC87606 similar to PIR|C84473|C84473 probable protein kinase [im... 223 3e-58
TC77160 similar to PIR|T48014|T48014 serine/threonine protein ki... 221 1e-57
TC87314 similar to PIR|T00848|T00848 probable serine/threonine-s... 219 5e-57
TC88193 similar to PIR|G96602|G96602 probable receptor protein k... 212 8e-57
TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein ki... 217 2e-56
TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenes... 216 2e-56
TC86201 similar to PIR|T49003|T49003 protein kinase-like protein... 216 3e-56
TC83862 similar to PIR|T05270|T05270 probable serine/threonine-s... 216 4e-56
TC79121 similar to GP|9651971|gb|AAF91337.1| Pti1 kinase-like pr... 211 8e-55
>TC88833 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 (EC 2.7.1.-)
receptor type precursor - Arabidopsis thaliana, partial
(36%)
Length = 1382
Score = 463 bits (1192), Expect = e-130
Identities = 226/346 (65%), Positives = 281/346 (80%), Gaps = 5/346 (1%)
Frame = +3
Query: 595 RGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCIN 654
+GGFG +Y+G L DGT+IA+KRM A+ KG +FQ+EI VL+KVRHRHLVALLG+C++
Sbjct: 48 QGGFGTIYEGGLHDGTRIALKRMMCGAIVGKGAAQFQSEIAVLTKVRHRHLVALLGYCLD 227
Query: 655 GNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIH 714
GNE+LLVYEYMPQGTL++++F W E G PL W +R+ +ALDVARGVEYLHSLA QSFIH
Sbjct: 228 GNEKLLVYEYMPQGTLSRYIFNWPEEGLEPLGWNKRLVIALDVARGVEYLHSLAHQSFIH 407
Query: 715 RDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVD 774
RDLKPSNILLGDDMRAKVADFGLV+ AP+GK S+ETR+AGTFGYLAPEYA TGRVTTKVD
Sbjct: 408 RDLKPSNILLGDDMRAKVADFGLVRLAPEGKASIETRIAGTFGYLAPEYAVTGRVTTKVD 587
Query: 775 VYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMES 834
V++FGV+LMELITGR+ALDDS P++ HLV WFRR+ ++K+ KAID T++ +EET+ S
Sbjct: 588 VFSFGVILMELITGRKALDDSQPEDSMHLVAWFRRMYLDKDTFRKAIDPTIDINEETLAS 767
Query: 835 IYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMSLPQVLQ 894
I+ V+ELAGHC+AREP QRPDMGHAVNVL +VEQWKP+ + +D + + +SLPQ L+
Sbjct: 768 IHTVAELAGHCSAREPYQRPDMGHAVNVLSSLVEQWKPSDTNAEDIYGIDLDLSLPQALK 947
Query: 895 RWQANEGTSTIFNDM-----SLSQTQSSINSKTYGFADSFDSLDCR 935
+WQA EG S + + SL TQ+SI ++ YGFADSF S D R
Sbjct: 948 KWQAYEGASQLDSSSSSLLPSLDNTQTSIPNRPYGFADSFTSADGR 1085
>TC76454 similar to PIR|T04124|T04124 receptor-like protein kinase (EC
2.7.1.-) - rice, partial (29%)
Length = 1269
Score = 384 bits (986), Expect(2) = e-112
Identities = 188/306 (61%), Positives = 236/306 (76%)
Frame = +3
Query: 630 FQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQ 689
F+ EI VLSKVRHRHLV L+G+ I GNER+LVYEYMPQG L+QHLF W+ G PL+WK+
Sbjct: 75 FRPEIAVLSKVRHRHLVGLIGYSIEGNERILVYEYMPQGALSQHLFHWKSFGLEPLSWKR 254
Query: 690 RVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVE 749
R+ +ALDVARG+EYLH+LA QSFIHRDLK SNILL DD RAKV+DFGLVK AP+G+ SV
Sbjct: 255 RLNIALDVARGMEYLHTLAHQSFIHRDLKSSNILLADDFRAKVSDFGLVKLAPNGEKSVV 434
Query: 750 TRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRR 809
T+LAGTFGYLAPEYA TG++TTKVDV++FGVVLMEL++G ALD+S P+E +L WF
Sbjct: 435 TKLAGTFGYLAPEYAVTGKITTKVDVFSFGVVLMELLSGMMALDESRPEESQYLAAWFWN 614
Query: 810 VLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQ 869
+ +K+ + AID TL+ +EET ES+ ++ELAGHCTAREPNQRP+MGHAVNVL P+VE+
Sbjct: 615 IKSDKKKLMAAIDPTLDINEETFESVSIIAELAGHCTAREPNQRPEMGHAVNVLAPLVEK 794
Query: 870 WKPTSRHEDDGHDSEPHMSLPQVLQRWQANEGTSTIFNDMSLSQTQSSINSKTYGFADSF 929
WKP D+ + + L Q+++ WQ EG T + M L ++SSI ++ GFADSF
Sbjct: 795 WKPFDDDPDEYSGIDYSLPLTQMVKGWQEAEGKDTSY--MDLEDSKSSIPARPTGFADSF 968
Query: 930 DSLDCR 935
S D R
Sbjct: 969 TSADGR 986
Score = 39.7 bits (91), Expect(2) = e-112
Identities = 19/27 (70%), Positives = 22/27 (81%)
Frame = +2
Query: 606 LQDGTKIAVKRMESVAMGNKGLNEFQA 632
L DGTKIAVKRME+ + NK L+EFQA
Sbjct: 2 LDDGTKIAVKRMEAGVITNKALDEFQA 82
>TC90987 similar to GP|15386483|emb|CAC60096. Seq ID: 318O13_region_A3~seq 4
513509 bases has been splitted in two~seq 4: 1 ->
349954 base~seq, partial (21%)
Length = 886
Score = 389 bits (998), Expect = e-108
Identities = 208/299 (69%), Positives = 228/299 (75%), Gaps = 6/299 (2%)
Frame = +2
Query: 302 QGPFPAFGKGVKVTLD-GINSFCKDTPGPCDARVMVLLHIAGAFGYPIKFAGSWKGNDPC 360
QGP P+FGK VKVTLD GINSFCK TPGPCD RV LL IA FGYP+ A SWKGNDPC
Sbjct: 2 QGPLPSFGKSVKVTLDEGINSFCKTTPGPCDPRVSTLLDIAAGFGYPLPLANSWKGNDPC 181
Query: 361 QGWSFVVCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLT 420
W+FVVC SG KIITVNLAKQ L GTIS AF NLTDLR+LYLNGNNLTGSIP SLT LT
Sbjct: 182 DDWTFVVC-SGGKIITVNLAKQNLNGTISSAFGNLTDLRNLYLNGNNLTGSIPGSLTGLT 358
Query: 421 QLETLEVSDNNLSGEVPKFPPKVKLLTAGNVLLGQTTGSGG-GAGGKTTPSAGSTPEGSH 479
QLE L+VS+NNLSGE+PKF KV+ +AGN LLG++ G GG G T PS G P GS
Sbjct: 359 QLEVLDVSNNNLSGEIPKFSGKVRFNSAGNGLLGKSEGDGGSGTAPPTDPSGG--PSGSP 532
Query: 480 GESGNGSSLTPGWIAGIVIIVLFFVAVVLFVSCKCYAK-RRHGKFSRVANPVNGNGNVKL 538
E G GSSL+PGWIAGI +I +FFVAVVLFV CKCYAK RRH KF RV NP NG +VK+
Sbjct: 533 PEKG-GSSLSPGWIAGIAVIAVFFVAVVLFVFCKCYAKNRRHTKFGRVNNPENGKNDVKI 709
Query: 539 DV---ISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILG 594
DV +S SNGY G PSELQSQ S L VF+GGN T+SI VL QVTGNF+EDNILG
Sbjct: 710 DVMSNVSNSNGYGGVPSELQSQGSERSDNLRVFEGGNVTISIQVLNQVTGNFNEDNILG 886
Score = 45.4 bits (106), Expect = 1e-04
Identities = 27/77 (35%), Positives = 37/77 (47%), Gaps = 1/77 (1%)
Frame = +2
Query: 243 QLAQVWLHKNQFTGPIPD-LSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNEL 301
++ V L K G I ++L +L L N LTG +P SL GLT L+ + + NN L
Sbjct: 215 KIITVNLAKQNLNGTISSAFGNLTDLRNLYLNGNNLTGSIPGSLTGLTQLEVLDVSNNNL 394
Query: 302 QGPFPAFGKGVKVTLDG 318
G P F V+ G
Sbjct: 395 SGEIPKFSGKVRFNSAG 445
>TC88699 weakly similar to GP|9293948|dbj|BAB01851.1 contains similarity to
receptor protein kinase~gene_id:MYM9.9 {Arabidopsis
thaliana}, partial (9%)
Length = 1219
Score = 158 bits (400), Expect(4) = 2e-82
Identities = 88/169 (52%), Positives = 113/169 (66%), Gaps = 2/169 (1%)
Frame = +2
Query: 226 NQD--NGLSGTIDVLSNMTQLAQVWLHKNQFTGPIPDLSQCSNLFDLQLRDNQLTGPVPN 283
NQD NG +GTI+V+S+M L+Q WL+ N FTG IP++S ++LFDLQL N L G VP+
Sbjct: 602 NQDRLNGFTGTIEVISSMRFLSQAWLNNNAFTGTIPNMSNSTHLFDLQLHSNGLIGLVPS 781
Query: 284 SLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDGINSFCKDTPGPCDARVMVLLHIAGA 343
SL L SL N+SLDNN L+GP P F K VK T + N+FC+ GPCD +VMV+L I A
Sbjct: 782 SLFSLPSLTNISLDNNNLEGPIPMFHKRVKATWES-NNFCRSNVGPCDPQVMVMLEIFAA 958
Query: 344 FGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQGLQGTISPAF 392
G+P +F+ KGND C F+ C G KI++V+ Q L G ISPAF
Sbjct: 959 LGHP-EFS-RIKGNDVCTDGVFLRCRRG-KIVSVDFRGQYLNGAISPAF 1096
Score = 117 bits (292), Expect(4) = 2e-82
Identities = 66/147 (44%), Positives = 89/147 (59%), Gaps = 2/147 (1%)
Frame = +1
Query: 81 SRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFA 140
S SL+GTLP+ LT + L NN+++G +PS+ L +L+T +LG NNFTS+P F
Sbjct: 181 SSSLSGTLPN-------LTYIDLHNNSLTGSLPSMFALFSLETIYLGHNNFTSIPGHCFQ 339
Query: 141 GLTDLQTLSLSDNPNLSPWTLP--TELTQSTNLITLELGTARLTGQLPESFFDKFPGLQS 198
L +QTL+LS+N NL PW P +L S + TL+L + G LP FD FP L +
Sbjct: 340 LLLGMQTLNLSNNLNLKPWLFPEAEDLGYSELMHTLDLEATNILGPLPSDVFDWFPRLHT 519
Query: 199 VRLSYNNLTGALPNSLAASAIENLWLN 225
V LS+NN+ G LP SL S + L LN
Sbjct: 520 VSLSHNNIRGTLPLSLGKSVVRYLRLN 600
Score = 52.4 bits (124), Expect(4) = 2e-82
Identities = 26/60 (43%), Positives = 36/60 (59%)
Frame = +3
Query: 15 LLFSITIIIAADDGPVFAATEDAAVMSKLLKSLSPPPSDWSSTTPFCQWDGIKCDSSNRV 74
LLFS+ ++ + A ++A MS+LLK+L+P PS WS+ T C W GI C SS V
Sbjct: 15 LLFSLLFVVTIN------AQDEANYMSQLLKALTPTPSGWSNNTHHCNWTGIVCQSSQVV 176
Score = 39.7 bits (91), Expect(4) = 2e-82
Identities = 19/32 (59%), Positives = 23/32 (71%)
Frame = +1
Query: 400 SLYLNGNNLTGSIPESLTTLTQLETLEVSDNN 431
+L L NN TGSIP+S TTL QL+ L+V NN
Sbjct: 1120 NLTLTNNNFTGSIPKSFTTLPQLQLLDVFRNN 1215
Score = 32.3 bits (72), Expect = 0.87
Identities = 21/58 (36%), Positives = 32/58 (54%)
Frame = +1
Query: 376 TVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLS 433
+ NL K ++S NLT + L+ N+LTGS+P S+ L LET+ + NN +
Sbjct: 154 SANLVKLWPSSSLSGTLPNLTYID---LHNNSLTGSLP-SMFALFSLETIYLGHNNFT 315
>TC80821 similar to GP|21239384|gb|AAM44275.1 receptor-like kinase RHG4
{Glycine max}, partial (16%)
Length = 728
Score = 263 bits (673), Expect = 2e-70
Identities = 123/148 (83%), Positives = 137/148 (92%)
Frame = +3
Query: 788 GRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTA 847
GRRALDD++PDERSHLV+WFRRVL+NKENIPKAIDQTLNPDEETMESIYK++ELAGHCTA
Sbjct: 3 GRRALDDTMPDERSHLVSWFRRVLVNKENIPKAIDQTLNPDEETMESIYKIAELAGHCTA 182
Query: 848 REPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMSLPQVLQRWQANEGTSTIFN 907
REP QRPDMGHAVNVLVP+VEQWKP++ E+DG+ + HMSLPQ LQRWQANEGTST+FN
Sbjct: 183 REPYQRPDMGHAVNVLVPLVEQWKPSNHEEEDGYGIDLHMSLPQALQRWQANEGTSTMFN 362
Query: 908 DMSLSQTQSSINSKTYGFADSFDSLDCR 935
DMS SQTQSSI SK GFADSFDS+DCR
Sbjct: 363 DMSFSQTQSSIPSKPSGFADSFDSMDCR 446
>BE204895 weakly similar to GP|15386544|em Seq ID: rhg4_Jack_amplicon
{Glycine max}, partial (19%)
Length = 609
Score = 237 bits (605), Expect = 1e-62
Identities = 124/209 (59%), Positives = 153/209 (72%)
Frame = +3
Query: 5 KTLLSLSTLSLLFSITIIIAADDGPVFAATEDAAVMSKLLKSLSPPPSDWSSTTPFCQWD 64
KTL+SLS L FS + A D D A MSKL KSL+PPPS WS + FC W+
Sbjct: 9 KTLISLSKLFFTFSFLLHTAIAD--------DGAFMSKLAKSLTPPPSGWSGNS-FCSWN 161
Query: 65 GIKCDSSNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTA 124
G+KCD S+RVT+++LAS+SLTGTLPSDLNSLSQLT+LSLQ+N+++G +PSLANL+ L+T
Sbjct: 162 GVKCDGSDRVTSLNLASKSLTGTLPSDLNSLSQLTTLSLQSNSLTGALPSLANLTMLQTV 341
Query: 125 FLGRNNFTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQ 184
FLG NNFTS+P F GLT LQ LSL++N NL PW LP + TQS+NL+ L+LG L G
Sbjct: 342 FLGGNNFTSIPDGCFVGLTSLQKLSLTENINLKPWKLPMDFTQSSNLVELDLGQTNLIGS 521
Query: 185 LPESFFDKFPGLQSVRLSYNNLTGALPNS 213
LP+ F LQ++RLSYNNLTG LPNS
Sbjct: 522 LPD-IFVPLVSLQNLRLSYNNLTGDLPNS 605
Score = 60.8 bits (146), Expect = 2e-09
Identities = 31/80 (38%), Positives = 48/80 (59%)
Frame = +3
Query: 354 WKGNDPCQGWSFVVCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIP 413
W GN C W+ V CD ++ ++NLA + L GT+ +L+ L +L L N+LTG++P
Sbjct: 132 WSGNSFCS-WNGVKCDGSDRVTSLNLASKSLTGTLPSDLNSLSQLTTLSLQSNSLTGALP 308
Query: 414 ESLTTLTQLETLEVSDNNLS 433
SL LT L+T+ + NN +
Sbjct: 309 -SLANLTMLQTVFLGGNNFT 365
Score = 60.8 bits (146), Expect = 2e-09
Identities = 47/142 (33%), Positives = 72/142 (50%), Gaps = 3/142 (2%)
Frame = +3
Query: 168 STNLITLELGTARLTGQLPESFFDKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQ 227
S + +L L + LTG LP S + L ++ L N+LTGALP+ + ++ ++L
Sbjct: 180 SDRVTSLNLASKSLTGTLP-SDLNSLSQLTTLSLQSNSLTGALPSLANLTMLQTVFLGG- 353
Query: 228 DNGLSGTIDVLSNMTQLAQVWLHKNQFTGP--IP-DLSQCSNLFDLQLRDNQLTGPVPNS 284
+N S +T L ++ L +N P +P D +Q SNL +L L L G +P+
Sbjct: 354 NNFTSIPDGCFVGLTSLQKLSLTENINLKPWKLPMDFTQSSNLVELDLGQTNLIGSLPDI 533
Query: 285 LMGLTSLQNVSLDNNELQGPFP 306
+ L SLQN+ L N L G P
Sbjct: 534 FVPLVSLQNLRLSYNNLTGDLP 599
>TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsis
thaliana}, partial (77%)
Length = 1571
Score = 228 bits (580), Expect = 1e-59
Identities = 123/291 (42%), Positives = 173/291 (59%), Gaps = 1/291 (0%)
Frame = +1
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
L T F E N++G GGFG V+KG L G +AVK++ G +G EF E+ +LS
Sbjct: 445 LATATRGFKEANLIGEGGFGKVFKGRLSTGELVAVKQLSHD--GRQGFQEFVTEVLMLSL 618
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVAR 699
+ H +LV L+G+C +G++RLLVYEYMP G+L HLF+ + PL+W R+ +A+ AR
Sbjct: 619 LHHSNLVKLIGYCTDGDQRLLVYEYMPMGSLEDHLFDLPQ-DKEPLSWSSRMKIAVGAAR 795
Query: 700 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYS-VETRLAGTFGY 758
G+EYLH A I+RDLK +NILL D K++DFGL K P G + V TR+ GT+GY
Sbjct: 796 GLEYLHCKADPPVIYRDLKSANILLDSDFSPKLSDFGLAKLGPVGDNTHVSTRVMGTYGY 975
Query: 759 LAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIP 818
APEYA +G++T K D+Y+FGVVL+ELITGRRA+D S +LV+W R ++
Sbjct: 976 CAPEYAMSGKLTLKSDIYSFGVVLLELITGRRAIDASKKPGEQNLVSWSRPYFSDRRKFV 1155
Query: 819 KAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQ 869
D L + +++ + C +P RP +G V L + Q
Sbjct: 1156HMADPLLQ-GHFPVRCLHQAIAITAMCLQEQPKFRPLIGDIVVALEYLASQ 1305
>TC86901 similar to GP|13937147|gb|AAK50067.1 AT5g13160/T19L5_120
{Arabidopsis thaliana}, partial (83%)
Length = 2028
Score = 227 bits (579), Expect = 1e-59
Identities = 140/357 (39%), Positives = 193/357 (53%), Gaps = 2/357 (0%)
Frame = +1
Query: 520 HGKFSRVANPVNGNGNVKLDVISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSILV 579
H SR+ + G KL S SNG S +LQ Q + T +
Sbjct: 334 HNNISRLPSSGPSGGVEKLR--STSNGSSKRELQLQLQLPAALKDGPPGQIAAQTFTFRE 507
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQD-GTKIAVKRMESVAMGNKGLNEFQAEITVLS 638
L T NF + LG GGFG VYKG L+ G +AVK+++ G +G EF E+ +LS
Sbjct: 508 LAAATKNFRPQSFLGEGGFGRVYKGRLETTGQAVAVKQLDR--NGLQGNREFLVEVLMLS 681
Query: 639 KVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVA 698
+ +LV+L+G+C +G++RLLVYE+MP G+L HL + PL W R+ +A A
Sbjct: 682 LLHSPNLVSLIGYCADGDQRLLVYEFMPLGSLEDHLHDL-PADKEPLDWNTRMKIAAGAA 858
Query: 699 RGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG-KYSVETRLAGTFG 757
+G+EYLH A I+RD K SNILL + K++DFGL K P G K V TR+ GT+G
Sbjct: 859 KGLEYLHDKANPPVIYRDFKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYG 1038
Query: 758 YLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENI 817
Y APEYA TG++T K DVY+FGVV +ELITGR+A+D + P +LVTW R + ++
Sbjct: 1039YCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSTRPHGEQNLVTWARPLFNDRRKF 1218
Query: 818 PKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTS 874
K D L M +Y+ +A C + RP +G V L + Q T+
Sbjct: 1219SKLADPRLQ-GRYPMRGLYQALAVASMCIQEQAAARPLIGDVVTALSYLANQAHDTN 1386
>TC77391 similar to GP|14719339|gb|AAK73157.1 putative protein kinase {Oryza
sativa}, partial (82%)
Length = 1753
Score = 226 bits (577), Expect = 2e-59
Identities = 134/340 (39%), Positives = 196/340 (57%), Gaps = 12/340 (3%)
Frame = +3
Query: 541 ISVSNGYSGAPSELQSQSSGDHSELHVFDGGN-STMSILVLRQVTGNFSEDNILGRGGFG 599
IS ++ S A + S+S G+ + N + S +R T NF D++LG GGFG
Sbjct: 384 ISSTSRNSSASISVTSRSEGE-----ILQSSNLKSFSYNEVRAATRNFRPDSVLGEGGFG 548
Query: 600 VVYKGELQD----------GTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALL 649
V+KG + + G +AVKR+ G++G E+ AEI L +++H +LV L+
Sbjct: 549 SVFKGWIDEHSHAATKPGMGIIVAVKRLNQE--GHQGHREWLAEINYLGQLQHPNLVKLI 722
Query: 650 GHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQ 709
G+C RLLVYE+MP+G++ HLF R + P +W R+ +AL A+G+ +LHS +
Sbjct: 723 GYCFEDEHRLLVYEFMPKGSMENHLFR-RGSYFQPFSWSLRMKIALGAAKGLAFLHS-TE 896
Query: 710 QSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG-KYSVETRLAGTFGYLAPEYAATGR 768
I+RD K SNILL + AK++DFGL ++ P G K V TR+ GT GY APEY ATG
Sbjct: 897 PKVIYRDFKTSNILLDSNYDAKLSDFGLARDGPTGDKSHVSTRVMGTRGYAAPEYLATGH 1076
Query: 769 VTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPD 828
+T K DVY+FGVVL+E+I+GRRA+D +LP +LV W + L NK + + +D L
Sbjct: 1077LTAKSDVYSFGVVLLEIISGRRAIDKNLPSGEHNLVEWAKPYLSNKRRVFRVMDPRLE-R 1253
Query: 829 EETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVE 868
+ + + + LA C + EP RP+M V L + E
Sbjct: 1254QYSHSRAHAAAALASQCLSVEPRIRPNMDEVVKTLEQLQE 1373
>TC79005 similar to GP|19423982|gb|AAL87287.1 putative protein kinase
{Arabidopsis thaliana}, partial (70%)
Length = 1723
Score = 226 bits (575), Expect = 4e-59
Identities = 136/340 (40%), Positives = 194/340 (57%), Gaps = 8/340 (2%)
Frame = +3
Query: 548 SGAPSELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQ 607
+G+ + + + +S +H+F LR T F D ILG GGFGVVYKG +
Sbjct: 279 TGSMNIKELREGAGYSNVHIFTYNE-------LRLATKQFRPDFILGEGGFGVVYKGVID 437
Query: 608 DG-------TKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLL 660
D T++A+K + G +G E+ AE+ L + H +LV L G+C RLL
Sbjct: 438 DSVRAGYNSTEVAIKELNRE--GFQGDREWLAEVNYLGQFSHPNLVKLFGYCCEDEHRLL 611
Query: 661 VYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPS 720
VYEYM +L +HLF R G T LTW +R+ +AL ARG+ +LH A++ I+RD K S
Sbjct: 612 VYEYMASDSLEKHLF--RRAGST-LTWSKRMKIALHAARGLAFLHG-AERPIIYRDFKTS 779
Query: 721 NILLGDDMRAKVADFGLVKNAPDGKYS-VETRLAGTFGYLAPEYAATGRVTTKVDVYAFG 779
NILL D AK++DFGL K+ P G + V TR+ GT+GY APEY TG +T + DVY FG
Sbjct: 780 NILLDADFNAKLSDFGLAKDGPMGDQTHVSTRVMGTYGYAAPEYVMTGHLTARSDVYGFG 959
Query: 780 VVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVS 839
VVL+EL+ GRRALD S P +LV W R +L + + + K +D + + + ++ KV+
Sbjct: 960 VVLLELLIGRRALDKSRPSREHNLVEWARPLLNHNKKLLKILDPKVE-GQYSSKTATKVA 1136
Query: 840 ELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDD 879
LA C ++ P RP M V +L E ++ +E+D
Sbjct: 1137LLAYQCLSQNPKSRPLMSQVVEIL----ENFQSKEENEED 1244
>TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC98010.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1347
Score = 224 bits (570), Expect = 2e-58
Identities = 127/317 (40%), Positives = 187/317 (58%), Gaps = 3/317 (0%)
Frame = +2
Query: 543 VSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVY 602
++NG S P L+S S ++ GG S + ++T FS +N++G GGFG VY
Sbjct: 77 MNNGGSIHP--LRSPSEATPPQM---SGGQILFSYDQILEITNGFSSENVIGEGGFGRVY 241
Query: 603 KGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVY 662
K + DG A+K ++ A +G EF+AE+ +S+V HRHLV+L+G+CI +R+L+Y
Sbjct: 242 KALMPDGRVGALKLLK--AGSGQGEREFRAEVDTISRVHHRHLVSLIGYCIAEQQRVLIY 415
Query: 663 EYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNI 722
E++P G L QHL E + + L W +R+ +A+ ARG+ YLH IHRD+K SNI
Sbjct: 416 EFVPNGNLDQHLHESQ---WNVLDWPKRMKIAIGAARGLAYLHEGCNPKIIHRDIKSSNI 586
Query: 723 LLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVL 782
LL D A+VADFGL + D V TR+ GTFGY+APEYA +G++T + DV++FGVVL
Sbjct: 587 LLDDSYEAQVADFGLARLTDDTNTHVSTRVMGTFGYMAPEYATSGKLTDRSDVFSFGVVL 766
Query: 783 MELITGRRALDDSLPDERSHLVTWFRRVL---INKENIPKAIDQTLNPDEETMESIYKVS 839
+EL+TGR+ +D + P LV W R +L I + + D L+ E ++++
Sbjct: 767 LELVTGRKPVDPTQPVGDESLVEWARPILLRAIETGDFSELADPRLHRQYIDSE-MFRMI 943
Query: 840 ELAGHCTAREPNQRPDM 856
E A C +RP M
Sbjct: 944 EAAAACIRHSAPKRPRM 994
>TC87606 similar to PIR|C84473|C84473 probable protein kinase [imported] -
Arabidopsis thaliana, partial (69%)
Length = 1803
Score = 223 bits (568), Expect = 3e-58
Identities = 127/321 (39%), Positives = 183/321 (56%), Gaps = 7/321 (2%)
Frame = +3
Query: 553 ELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKI 612
EL + ++ + + + ++ L+ +T FS N LG GGFG V+KG + D +
Sbjct: 387 ELSNPNATEDLSISLAGSNLYAFTVAELKVITQQFSSSNHLGAGGFGPVHKGFIDDKVRP 566
Query: 613 AVKRMESVAM------GNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMP 666
+K +SVA+ +G E+ E+ VL ++R HLV L+G+CI RLLVYEY+P
Sbjct: 567 GLKA-QSVAVKLLDLESKQGHKEWLTEVVVLGQLRDPHLVKLIGYCIEDEHRLLVYEYLP 743
Query: 667 QGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGD 726
+G+L LF L W R+ +A+ A+G+ +LH A+Q I RD K SNILL
Sbjct: 744 RGSLENQLFRRFSAS---LPWSTRMKIAVGAAKGLAFLHE-AEQPVIFRDFKASNILLDS 911
Query: 727 DMRAKVADFGLVKNAPDGKYS-VETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMEL 785
D AK++DFGL K+ P+G + V TR+ GT GY APEY TG +T K DVY+FGVVL+EL
Sbjct: 912 DYNAKLSDFGLAKDGPEGDDTHVSTRVMGTQGYAAPEYVMTGHLTAKSDVYSFGVVLLEL 1091
Query: 786 ITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHC 845
+TGR+++D + P +LV W R +LI+ I K +D L M K + LA C
Sbjct: 1092LTGRKSVDKNRPQREQNLVDWARPMLIDSRKISKIMDPKLEGQYSEM-GAKKAASLAYQC 1268
Query: 846 TAREPNQRPDMGHAVNVLVPM 866
+ P RP M + V +L P+
Sbjct: 1269LSHRPKSRPTMSNVVKILEPL 1331
>TC77160 similar to PIR|T48014|T48014 serine/threonine protein kinase-like
protein - Arabidopsis thaliana, partial (94%)
Length = 1493
Score = 221 bits (562), Expect = 1e-57
Identities = 120/293 (40%), Positives = 186/293 (62%), Gaps = 5/293 (1%)
Frame = +3
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
L++VT NF +D+++G G +G VY G L+ G A+K++++ ++ EF A+++++S+
Sbjct: 381 LKEVTDNFGQDSLIGEGSYGRVYYGVLKSGQAAAIKKLDASKQPDE---EFLAQVSMVSR 551
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWREL-GYTP---LTWKQRVTVAL 695
++H + V LLG+C++GN R+L YE+ G+L L + + G P LTW QRV +A+
Sbjct: 552 LKHDNFVQLLGYCVDGNSRILAYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAV 731
Query: 696 DVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVE-TRLAG 754
ARG+EYLH A IHRD+K SN+L+ DD AK+ADF L APD + TR+ G
Sbjct: 732 GAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLG 911
Query: 755 TFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINK 814
TFGY APEYA TG++ K DVY+FGVVL+EL+TGR+ +D +LP + LVTW L ++
Sbjct: 912 TFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKL-SE 1088
Query: 815 ENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMV 867
+ + + +D L E +++ K++ +A C E + RP+M V L P++
Sbjct: 1089DKVRQCVDTRLG-GEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLL 1244
>TC87314 similar to PIR|T00848|T00848 probable serine/threonine-specific
protein kinase T20F6.6 (EC 2.7.1.-) - Arabidopsis
thaliana, partial (80%)
Length = 1575
Score = 219 bits (557), Expect = 5e-57
Identities = 140/376 (37%), Positives = 201/376 (53%), Gaps = 27/376 (7%)
Frame = +1
Query: 533 NGNVKLDVISVSNGYSGA----PSELQSQSSGD-----------HSELHVFDGGN-STMS 576
+ + K+D S SG PS L S D SE + N S
Sbjct: 223 DSSAKVDAAQSSRSTSGTSKTTPSSLTIPSYSDKSNSSSLLPTPRSEGEILSSPNLKAFS 402
Query: 577 ILVLRQVTGNFSEDNILGRGGFGVVYKGELQD----------GTKIAVKRMESVAMGNKG 626
L+ T NF D++LG GGFG VYKG + + G +AVKR++ G +G
Sbjct: 403 FNELKNATRNFRPDSLLGEGGFGHVYKGWIDEHTFTAAKPGSGMVVAVKRLKPE--GYQG 576
Query: 627 LNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLT 686
E+ E+ L ++ H +LV L+G+C+ G RLLVYE+MP+G+L HLF G PL+
Sbjct: 577 HKEWLTEVNYLGQLHHPNLVKLIGYCLEGENRLLVYEFMPKGSLENHLFR---RGPQPLS 747
Query: 687 WKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG-K 745
W R+ VA+ ARG+ +LH+ A+ I+RD K SNILL + +K++DFGL K P G +
Sbjct: 748 WSIRMKVAIGAARGLSFLHN-AKSQVIYRDFKASNILLDAEFNSKLSDFGLAKAGPTGDR 924
Query: 746 YSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVT 805
V T++ GT GY APEY ATGR+T K DVY+FGVV++EL++GRRA+D ++ +LV
Sbjct: 925 THVSTQVVGTQGYAAPEYVATGRLTAKSDVYSFGVVMLELLSGRRAVDKTIAGVDQNLVD 1104
Query: 806 WFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVP 865
W + L +K + + +D L + + + + LA C RE RP M + L
Sbjct: 1105WAKPYLGDKRRLFRIMDSKLE-GQYPQKGAFMAATLALQCLNREAKARPSMTEVLATL-E 1278
Query: 866 MVEQWKPTSRHEDDGH 881
+E K SR+ H
Sbjct: 1279QIEAPKHASRNSLSEH 1326
>TC88193 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(20%)
Length = 1815
Score = 212 bits (540), Expect(2) = 8e-57
Identities = 114/246 (46%), Positives = 156/246 (63%)
Frame = +3
Query: 561 DHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESV 620
D +L D +T S L+ T +F+ DN LG GGFG VYKG L DG +AVK++
Sbjct: 498 DDDDLVGIDTMPNTFSYYELKNATSDFNRDNKLGEGGFGPVYKGTLNDGRFVAVKQLSIG 677
Query: 621 AMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWREL 680
+ ++G ++F AEI +S V+HR+LV L G CI GN+RLLVYEY+ +L Q LF
Sbjct: 678 S--HQGKSQFIAEIATISAVQHRNLVKLYGCCIEGNKRLLVYEYLENKSLDQALFG---- 839
Query: 681 GYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKN 740
L W R V + VARG+ YLH ++ +HRD+K SNILL ++ K++DFGL K
Sbjct: 840 NVLFLNWSTRYDVCMGVARGLTYLHEESRLRIVHRDVKASNILLDSELVPKLSDFGLAKL 1019
Query: 741 APDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDER 800
D K + TR+AGT GYLAPEYA GR+T K DV++FGVV +EL++GR D SL +++
Sbjct: 1020YDDKKTHISTRVAGTIGYLAPEYAMRGRLTEKADVFSFGVVALELVSGRPNSDSSLEEDK 1199
Query: 801 SHLVTW 806
+L+ W
Sbjct: 1200MYLLDW 1217
Score = 27.7 bits (60), Expect(2) = 8e-57
Identities = 14/33 (42%), Positives = 17/33 (51%)
Frame = +1
Query: 845 CTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHE 877
CT PN RP M V +L+ +E TSR E
Sbjct: 1324 CTQTSPNLRPSMSRVVAMLLGDIEVSTVTSRPE 1422
>TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein kinase
{Arabidopsis thaliana}, partial (72%)
Length = 2379
Score = 217 bits (552), Expect = 2e-56
Identities = 119/290 (41%), Positives = 173/290 (59%), Gaps = 6/290 (2%)
Frame = +2
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGL--NEFQAEITVL 637
L T FS+DNI+G GG+GVVY+G+L +G +A+K++ + N G EF+ E+ +
Sbjct: 704 LELATNKFSKDNIIGEGGYGVVYQGQLINGNPVAIKKL----LNNLGQAEKEFRVEVEAI 871
Query: 638 SKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLF-EWRELGYTPLTWKQRVTVALD 696
VRH++LV LLG CI G RLL+YEY+ G L Q L R+ GY LTW R+ + L
Sbjct: 872 GHVRHKNLVRLLGFCIEGTHRLLIYEYVNNGNLEQWLHGAMRQYGY--LTWDARIKILLG 1045
Query: 697 VARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTF 756
A+ + YLH + +HRD+K SNIL+ DD AK++DFGL K GK + TR+ GTF
Sbjct: 1046TAKALAYLHEAIEPKVVHRDIKSSNILIDDDFNAKISDFGLAKLLGAGKSHITTRVMGTF 1225
Query: 757 GYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKEN 816
GY+APEYA +G + K DVY+FGV+L+E ITGR +D + +LV W + ++ N+
Sbjct: 1226GYVAPEYANSGLLNEKSDVYSFGVLLLEAITGRDPVDYNRSAAEVNLVDWLKMMVGNRH- 1402
Query: 817 IPKAIDQTLNPDEETMES---IYKVSELAGHCTAREPNQRPDMGHAVNVL 863
++ ++P+ ET S + +V A C + +RP M V +L
Sbjct: 1403----AEEVVDPNIETRPSTSALKRVLLTALRCVDPDSEKRPKMSQVVRML 1540
>TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenesis
receptor-like kinase putative {Arabidopsis thaliana},
partial (77%)
Length = 1762
Score = 216 bits (551), Expect = 2e-56
Identities = 119/288 (41%), Positives = 170/288 (58%)
Frame = +1
Query: 576 SILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEIT 635
S+ L T NF+ DN LG GGFG VY G+L DG++IAVKR++ NK EF E+
Sbjct: 430 SLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLK--VWSNKADMEFAVEVE 603
Query: 636 VLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVAL 695
+L++VRH++L++L G+C G ERL+VY+YMP +L HL + + L W +R+ +A+
Sbjct: 604 ILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHL-HGQHSTESLLDWNRRMNIAI 780
Query: 696 DVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGT 755
A G+ YLH A IHRD+K SN+LL D +A+VADFG K PDG V TR+ GT
Sbjct: 781 GSAEGIVYLHVQATPHIIHRDVKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGT 960
Query: 756 FGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKE 815
GYLAPEYA G+ DVY+FG++L+EL +G++ L+ + + W + K+
Sbjct: 961 LGYLAPEYAMLGKANESCDVYSFGILLLELASGKKPLEKLSSSVKRAINDWALPLACEKK 1140
Query: 816 NIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVL 863
+ D LN D E + +V +A C +P +RP M V +L
Sbjct: 1141-FSELADPRLNGD-YVEEELKRVILVALICAQNQPEKRPTMVEVVELL 1278
>TC86201 similar to PIR|T49003|T49003 protein kinase-like protein -
Arabidopsis thaliana, partial (86%)
Length = 2127
Score = 216 bits (550), Expect = 3e-56
Identities = 119/303 (39%), Positives = 186/303 (61%), Gaps = 5/303 (1%)
Frame = +3
Query: 575 MSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEI 634
+S+ L++ T NF ++G G +G VY L DG +AVK+++ V+ + NEF ++
Sbjct: 378 LSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGKAVAVKKLD-VSTEPESNNEFLTQV 554
Query: 635 TVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWREL-GYTP---LTWKQR 690
+++S++++ + V L G+C+ GN R+L YE+ G+L L + + G P L W QR
Sbjct: 555 SMVSRLKNENFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLNWMQR 734
Query: 691 VTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVE- 749
V +A+D ARG+EYLH Q S IHRD++ SN+L+ +D +AKVADF L APD +
Sbjct: 735 VRIAVDAARGLEYLHEKVQPSIIHRDIRSSNVLIFEDYKAKVADFNLSNQAPDMAARLHS 914
Query: 750 TRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRR 809
TR+ GTFGY APEYA TG++T K DVY+FGVVL+EL+TGR+ +D ++P + LVTW
Sbjct: 915 TRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTW-AT 1091
Query: 810 VLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQ 869
++++ + + +D L E + + K++ +A C E RP+M V L P+++
Sbjct: 1092PRLSEDKVKQCVDPKLK-GEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPLLKA 1268
Query: 870 WKP 872
P
Sbjct: 1269PAP 1277
>TC83862 similar to PIR|T05270|T05270 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) T4L20.80 - Arabidopsis
thaliana, partial (60%)
Length = 948
Score = 216 bits (549), Expect = 4e-56
Identities = 117/291 (40%), Positives = 178/291 (60%), Gaps = 3/291 (1%)
Frame = +1
Query: 576 SILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGL--NEFQAE 633
S+ L T F+E +++G G+G+VY+G LQDG+ +AVK + + NKG EF+ E
Sbjct: 16 SLKELENATDGFAEGSVIGERGYGIVYRGILQDGSIVAVKNL----LNNKGQAEKEFKVE 183
Query: 634 ITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELG-YTPLTWKQRVT 692
+ + KVRH++LV L+G+C G +R+LVYEY+ G L Q L ++G +PLTW R+
Sbjct: 184 VEAIGKVRHKNLVGLVGYCAEGAKRMLVYEYVDNGNLEQWLHG--DVGPVSPLTWDIRMK 357
Query: 693 VALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRL 752
+A+ A+G+ YLH + +HRD+K SNILL AKV+DFGL K GK V TR+
Sbjct: 358 IAVGTAKGLAYLHEGLEPKVVHRDVKSSNILLDKKWHAKVSDFGLAKLLGSGKSYVTTRV 537
Query: 753 AGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLI 812
GTFGY++PEYA+TG + DVY+FG++LMEL+TGR +D S +LV WF+ ++
Sbjct: 538 MGTFGYVSPEYASTGMLNEGSDVYSFGILLMELVTGRSPIDYSRAPAEMNLVDWFKGMVA 717
Query: 813 NKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVL 863
++ + + L + + S+ + + C + N+RP MG V++L
Sbjct: 718 SRRG--EELVDPLIEIQPSPRSLKRALLVCLRCIDLDANKRPKMGQIVHML 864
>TC79121 similar to GP|9651971|gb|AAF91337.1| Pti1 kinase-like protein
{Glycine max}, partial (96%)
Length = 1283
Score = 211 bits (538), Expect = 8e-55
Identities = 115/303 (37%), Positives = 186/303 (60%), Gaps = 5/303 (1%)
Frame = +3
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
LR +T NF +G G +G VY+ L++G ++A+K+++S ++ EF ++++++S+
Sbjct: 324 LRSLTDNFGTKTFVGEGAYGKVYRATLKNGREVAIKKLDSSKQPDQ---EFLSQVSIVSR 494
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWREL-GYTP---LTWKQRVTVAL 695
++H ++V L+ +C++G R L YEY P G+L L + + G P L+W +RV +A+
Sbjct: 495 LKHENVVELVNYCVDGPLRALAYEYAPNGSLHDILHGRKGVKGAEPGQVLSWAERVKIAV 674
Query: 696 DVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVE-TRLAG 754
ARG+EYLH A+ +HR +K SNILL +D AK+ADF L APD + TR+ G
Sbjct: 675 GAARGLEYLHEKAEVHIVHRYIKSSNILLFEDGVAKIADFDLSNQAPDAAARLHSTRVLG 854
Query: 755 TFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINK 814
TFGY APEYA TG +++K DVY+FGV+L+EL+TGR+ +D +LP + LVTW L ++
Sbjct: 855 TFGYHAPEYAMTGNLSSKSDVYSFGVILLELLTGRKPVDHTLPRGQQSLVTWATPKL-SE 1031
Query: 815 ENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTS 874
+ + + +D L E +S+ K++ +A C E RP+M V L P++ + +
Sbjct: 1032DKVKQCVDVRLK-GEYPSKSVAKLAAVAALCVQYEAEFRPNMSIIVKALQPLMNNTRSSQ 1208
Query: 875 RHE 877
E
Sbjct: 1209PRE 1217
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,899,842
Number of Sequences: 36976
Number of extensions: 392832
Number of successful extensions: 5083
Number of sequences better than 10.0: 912
Number of HSP's better than 10.0 without gapping: 3302
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4089
length of query: 935
length of database: 9,014,727
effective HSP length: 105
effective length of query: 830
effective length of database: 5,132,247
effective search space: 4259765010
effective search space used: 4259765010
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0108.8


