

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
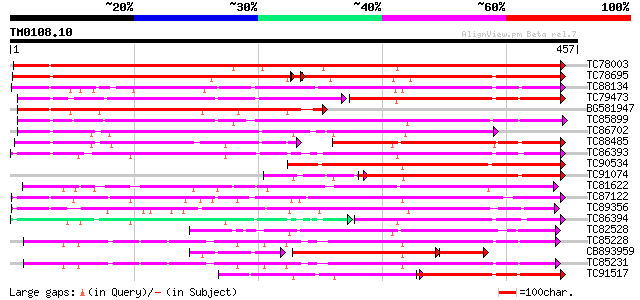
Query= TM0108.10
(457 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase... 596 e-171
TC78695 similar to GP|4115563|dbj|BAA36423.1 UDP-glucose:anthocy... 284 e-143
TC88134 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 295 3e-80
TC79473 weakly similar to PIR|T00639|T00639 hypothetical protein... 163 7e-79
BG581947 similar to GP|20146093|dbj glucosyltransferase NTGT2 {N... 277 7e-75
TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:... 276 1e-74
TC86702 weakly similar to GP|9794913|gb|AAF98390.1| UDP-glucose:... 231 3e-61
TC88485 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 159 1e-54
TC86393 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyl... 193 1e-49
TC90534 weakly similar to GP|19911197|dbj|BAB86925. glucosyltran... 186 1e-47
TC91074 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 152 1e-43
TC81622 weakly similar to PIR|T00584|T00584 indole-3-acetate bet... 172 3e-43
TC87122 weakly similar to GP|19911203|dbj|BAB86928. glucosyltran... 171 7e-43
TC89356 similar to GP|19911207|dbj|BAB86930. glucosyltransferase... 169 3e-42
TC86394 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyl... 120 7e-42
TC82528 weakly similar to PIR|C86356|C86356 hypothetical protein... 164 5e-41
TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 162 3e-40
CB893959 weakly similar to GP|7385017|gb| UDP-glucose:salicylic ... 103 4e-40
TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {R... 160 7e-40
TC91517 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 115 5e-39
>TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase NTGT2
{Nicotiana tabacum}, partial (50%)
Length = 1740
Score = 596 bits (1537), Expect = e-171
Identities = 302/459 (65%), Positives = 358/459 (77%), Gaps = 14/459 (3%)
Frame = +3
Query: 4 HRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPFSD 63
HR LL+ YP QGHINP +FAKRLI+ GA HVT+ TT+H + RI+NKPTLPNL+ PFSD
Sbjct: 147 HRILLVPYPVQGHINPAFEFAKRLITLGA-HVTISTTVHMHNRITNKPTLPNLSYYPFSD 323
Query: 64 GFDDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWAAEAAR 123
G+DDGFK T Y Y E +RRG+EFV+D+IL +QEG PFTCL+++LLL WAAEAAR
Sbjct: 324 GYDDGFKGTGSDAYLEYHAEFQRRGSEFVSDIILKNSQEGTPFTCLVHSLLLQWAAEAAR 503
Query: 124 GLHLPTALLWVQPATVLDIIYYYFHGYHDCLLS-KIEFELPGLPFSLSPSDLPSFLL--- 179
HLPTALLWVQPATV DI+YYYFHG+ D + + ELPGLP S DLPSFLL
Sbjct: 504 EFHLPTALLWVQPATVFDILYYYFHGFSDSIKNPSSSIELPGLPLLFSSRDLPSFLLASC 683
Query: 180 PSNDSFIVSYFEEQFRELDVEAN--PTILVNSFEALEPEAMRAVEEFNVIPIGPLIPSAF 237
P S + S+FEEQF ELDVE N TILVNSFE+LEP+A+RAV++FN+I IGPLIPS
Sbjct: 684 PDAYSLMTSFFEEQFNELDVETNLTKTILVNSFESLEPKALRAVKKFNMISIGPLIPSEH 863
Query: 238 LDGKD-LNDTSFGGD--IFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARAL 294
LD KD D S+GG IF+ SN C EWLD + + SVVYVSFGS+ VLS+ Q EEIA AL
Sbjct: 864 LDEKDSTEDNSYGGQTHIFQPSNDCVEWLDSKPKSSVVYVSFGSYFVLSERQREEIAHAL 1043
Query: 295 LDCGHPFLWVIREMEEE-----LSCREELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWN 349
LDCG PFLWV+RE E E REELEEKGK+V WCSQ+E+LSH S+GCFLTHCGWN
Sbjct: 1044LDCGFPFLWVLREKEGENNEEGFKYREELEEKGKIVKWCSQMEILSHPSLGCFLTHCGWN 1223
Query: 350 STMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAV 409
ST+ESLV GVPMVAFP+++DQMTNAK++EDVWKIGVR+D VNEDG+V G EIR+CL+ V
Sbjct: 1224STLESLVKGVPMVAFPQWTDQMTNAKLIEDVWKIGVRVDEEVNEDGIVRGDEIRRCLEVV 1403
Query: 410 MGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
MG+GEK EE+RR+ +K+K LA EA KEGGSSEKNL +FL
Sbjct: 1404MGSGEKGEELRRSGKKWKELAREAVKEGGSSEKNLRSFL 1520
>TC78695 similar to GP|4115563|dbj|BAA36423.1 UDP-glucose:anthocyanin
5-O-glucosyltransferase {Verbena x hybrida}, partial
(49%)
Length = 1741
Score = 284 bits (727), Expect(3) = e-143
Identities = 145/227 (63%), Positives = 176/227 (76%), Gaps = 15/227 (6%)
Frame = +1
Query: 237 FLDGKDLNDTSFGGDIFRLSN--GCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARAL 294
FLDGKD D SFGGD+ R+ + +WLD + EKSVVYVSFG+ VLSK QMEEIARAL
Sbjct: 802 FLDGKDPTDNSFGGDVVRVDSKDDYIQWLDSKDEKSVVYVSFGTLAVLSKRQMEEIARAL 981
Query: 295 LDCGHPFLWVIREM-----------EEELSCREELEEK--GKVVTWCSQVEVLSHRSVGC 341
LD G FLWVIR+ ++ELSCREELE GK+V WCSQVEVLSHRS+GC
Sbjct: 982 LDSGFSFLWVIRDKKLQQQKEEEVDDDELSCREELENNMNGKIVKWCSQVEVLSHRSLGC 1161
Query: 342 FLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGE 401
F+THCGWNST+ESL SGVPMVAFP+++DQ TNAK++EDVWK G+RM+H +E+G+V+ E
Sbjct: 1162 FMTHCGWNSTLESLGSGVPMVAFPQWTDQTTNAKLIEDVWKTGLRMEH--DEEGMVKVEE 1335
Query: 402 IRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
IRKCL+ VMG GEK EE+RRNA+K+K LA A KEGGSS +NL ++L
Sbjct: 1336 IRKCLEVVMGKGEKGEELRRNAKKWKDLARAAVKEGGSSNRNLRSYL 1476
Score = 239 bits (611), Expect(3) = e-143
Identities = 126/236 (53%), Positives = 160/236 (67%), Gaps = 8/236 (3%)
Frame = +2
Query: 3 HHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPFS 62
+H FL+I YP QGHINP LQF KRLIS GA+ VT TT+H Y R+ NKPT+P L+ FS
Sbjct: 77 NHHFLIITYPLQGHINPALQFTKRLISLGAK-VTFATTIHLYSRLINKPTIPGLSFATFS 253
Query: 63 DGFDDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWAAEAA 122
DG+DDG K D D Y +E RRG+EF+T++ILS+ QE PFTCLIYTL+L WA + A
Sbjct: 254 DGYDDGQKSFGDEDIVSYMSEFTRRGSEFLTNIILSSKQENHPFTCLIYTLILSWAPKVA 433
Query: 123 RGLHLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFE-----LPGLPFSLSPSDLPSF 177
LHLP+ LLW+Q ATV DI YYYFH + D + +K + E LPGL FSL DLPSF
Sbjct: 434 HELHLPSTLLWIQAATVFDIFYYYFHEHGDYITNKSKDETCLISLPGLSFSLKSRDLPSF 613
Query: 178 LLPSND-SFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVE--EFNVIPIG 230
LL SN +F + +EQ + L+ E NP +LVN+ E E +A+ V+ + +IPIG
Sbjct: 614 LLASNTYTFALPSLKEQIQLLNEEINPRVLVNTVEEFELDALNKVDVGKIKMIPIG 781
Score = 24.3 bits (51), Expect(3) = e-143
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = +3
Query: 228 PIGPLIPSAFL 238
P GPLIPSAFL
Sbjct: 774 PSGPLIPSAFL 806
>TC88134 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (27%)
Length = 1669
Score = 295 bits (754), Expect = 3e-80
Identities = 181/474 (38%), Positives = 277/474 (58%), Gaps = 27/474 (5%)
Frame = +2
Query: 2 PHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRI-----SNKPTLPN- 55
P LL+ + AQGHINP L+ K L++ G +VTL TT Y R+ ++ T+P
Sbjct: 65 PEIHVLLVAFSAQGHINPLLRLGKSLLTKGL-NVTLATTELVYHRVFKSTTTDTDTVPTS 241
Query: 56 -----LTLLPFSDGFD--DGFKLTPDTDYSLYATELKRRGTEFVTDLILS--AAQEGKPF 106
+ +L FSDGFD G K +PD L A + G +T+LI + K
Sbjct: 242 YSTNGINVLFFSDGFDISQGHK-SPDEYMELIA----KFGPISLTNLINNNFINNPSKKL 406
Query: 107 TCLIYTLLLHWAAEAARGLHLPTALLWVQPATVLDIIYYYFHGYHDCLL---SKIEFELP 163
C+I + W A A L++P A LW+QP T+ I Y +++ + +I+ ELP
Sbjct: 407 ACIINNPFVPWVANVAYELNIPCACLWIQPCTLYSIYYRFYNELNQFPTLENPEIDVELP 586
Query: 164 GLPFSLSPSDLPSFLLPSNDSFIVS-YFEEQFRELDVEANPTILVNSFEALEPEAMRAVE 222
GLP L P DLPSF+LPSN +S E F+ D++ +L NSF LE + + ++
Sbjct: 587 GLPL-LKPQDLPSFILPSNPIKTMSDVLAEMFQ--DMKKLKWVLANSFYELEKDVIDSMA 757
Query: 223 E-FNVIPIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVV 281
E F + P+GPL+P + L D G +++ + C EWL+ + SV+Y+SFGS +
Sbjct: 758 EIFPITPVGPLVPPSLLGQDQKQDV--GIEMWTPQDSCMEWLNQKLPSSVIYISFGSLIF 931
Query: 282 LSKTQMEEIARALLDCGHPFLWVIREME-----EELSCR--EELEEKGKVVTWCSQVEVL 334
LS+ QM+ IA AL + FLWV++ + +LS + EE +EKG VVTWC Q +VL
Sbjct: 932 LSEKQMQSIASALKNSNKYFLWVMKSNDIGAEKVQLSEKFLEETKEKGLVVTWCPQTKVL 1111
Query: 335 SHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNED 394
H ++ CFLTHCGWNST+E++ +GVPM+ +P+++DQ TNA +V DV+++G+R+ + D
Sbjct: 1112VHPAIACFLTHCGWNSTLEAITAGVPMIGYPQWTDQPTNATLVSDVFRMGIRLKQ--DND 1285
Query: 395 GVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
G VE E+ + ++ ++G GE++E ++NA + K A EA +GGSS++N+ F+
Sbjct: 1286GFVESDEVERAIEEIVG-GERSEVFKKNALELKHAAREAVADGGSSDRNIQRFV 1444
>TC79473 weakly similar to PIR|T00639|T00639 hypothetical protein F3I6.2 -
Arabidopsis thaliana, partial (45%)
Length = 1530
Score = 163 bits (413), Expect(2) = 7e-79
Identities = 83/177 (46%), Positives = 124/177 (69%), Gaps = 3/177 (1%)
Frame = +1
Query: 275 SFGSFVVLSKTQMEEIARALLDCGHPFLWVIREMEE---ELSCREELEEKGKVVTWCSQV 331
SFGS V L+ Q+EE+A L + FLWV+RE E+ ++ ++EKG +VTWC+Q+
Sbjct: 868 SFGSMVSLTSEQIEELALGLKESEVNFLWVLRESEQGKLPKGYKDFIKEKGIIVTWCNQL 1047
Query: 332 EVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSV 391
E+L+H +VGCF+THCGWNST+ESL GVP+V P+++DQ+ +AK +E++W++GVR
Sbjct: 1048 ELLAHDAVGCFVTHCGWNSTLESLSLGVPVVCLPQWADQLPDAKFLEEIWEVGVRPKE-- 1221
Query: 392 NEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
+E+GVV+ E L VM E++E +RRNA ++K LA +A EGGS +KN+ F+
Sbjct: 1222 DENGVVKREEFMLSLKVVM-ESERSEVIRRNASEWKKLARDAVCEGGSFDKNINQFV 1389
Score = 149 bits (375), Expect(2) = 7e-79
Identities = 94/269 (34%), Positives = 150/269 (54%), Gaps = 4/269 (1%)
Frame = +2
Query: 7 LLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPFSDGFD 66
L+I YPAQGHI+P +QF+KRL+S G + T TT +T + I T PN+++ P SDGFD
Sbjct: 80 LVIPYPAQGHISPLIQFSKRLVSKGIK-TTFATTHYTVKSI----TAPNISVEPISDGFD 244
Query: 67 D-GFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWAAEAARGL 125
+ GF + + L+ K G++ +++LI + P TC++Y L WA + A+
Sbjct: 245 ESGFSQAKNVE--LFLNSFKTNGSKTLSNLIQKHQKTSTPITCIVYDSFLPWALDVAKQH 418
Query: 126 HLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFELPGLPFSLSPSDLPSFL-LPSNDS 184
+ A + A V +I HG + + ++ +PGLP L+ DLPSF+ P +
Sbjct: 419 RIYGAAFFTNSAAVCNIFCRIHHGLIETPVDELPLIVPGLP-PLNSRDLPSFIRFPESYP 595
Query: 185 FIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVEE-FNVIPIGPLIPSAFLDGKDL 243
++ QF L+ + VN+FEALE E ++ + E F IGP++PSA+LDG+
Sbjct: 596 AYMAMKLNQFSNLN--QADWMFVNTFEALEAEVVKGLTEMFPAKLIGPMVPSAYLDGRIK 769
Query: 244 NDTSFGGDIFR-LSNGCAEWLDLQAEKSV 271
D +G ++++ LS C WL+ + +SV
Sbjct: 770 GDKGYGANLWKPLSEDCINWLNAKPSQSV 856
>BG581947 similar to GP|20146093|dbj glucosyltransferase NTGT2 {Nicotiana
tabacum}, partial (8%)
Length = 786
Score = 277 bits (708), Expect = 7e-75
Identities = 147/268 (54%), Positives = 186/268 (68%), Gaps = 18/268 (6%)
Frame = +2
Query: 7 LLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRI----SNKPTLPNLTLLPFS 62
LL+ YPAQGHINP LQFAKRLIS GA HVTL TLH YRR+ + T+ NL++ PFS
Sbjct: 5 LLVTYPAQGHINPALQFAKRLISMGA-HVTLPITLHLYRRLILLNPSITTISNLSITPFS 181
Query: 63 DGFDDGFKL--TPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWAAE 120
DG++DGF D D+ Y ++ RG++F+T+LILSA QE KPFTCL+YT+++ WA
Sbjct: 182 DGYNDGFIAITNTDADFHQYTSQFNTRGSDFITNLILSAKQESKPFTCLLYTIIIPWAPR 361
Query: 121 AARGLHLPTALLWVQPATVLDIIYYYFHGYHDCL------LSKIEFELPGLPFSLSPSDL 174
ARG +L +A LW++PATV DI+YYYFHGY + + ++ ELPGLPF+LSP D+
Sbjct: 362 VARGFNLRSAKLWIEPATVFDILYYYFHGYSNHINNQNQNQNQTTIELPGLPFTLSPRDI 541
Query: 175 PSFLLPSND---SFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVE---EFNVIP 228
PSFL SN SF+ YF++ F ELDVE NP ILVN+FEALEPEA+RAV+ +IP
Sbjct: 542 PSFLFTSNPSVLSFVFPYFQQDFHELDVETNPIILVNTFEALEPEALRAVDTHHNLKMIP 721
Query: 229 IGPLIPSAFLDGKDLNDTSFGGDIFRLS 256
IGPLIPS DTSF GD+ + S
Sbjct: 722 IGPLIPS---------DTSFSGDLLQPS 778
>TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:salicylic
acid glucosyltransferase {Nicotiana tabacum}, partial
(47%)
Length = 1617
Score = 276 bits (706), Expect = 1e-74
Identities = 165/452 (36%), Positives = 259/452 (56%), Gaps = 9/452 (1%)
Frame = +2
Query: 7 LLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPFSDGFD 66
L++ YPAQGH+NP +QF+KRLI G + +TL T ++ ISNK L ++ + SDG+D
Sbjct: 50 LILPYPAQGHMNPMIQFSKRLIEKGVK-ITLITVTSFWKVISNK-NLTSIDVESISDGYD 223
Query: 67 DGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWAAEAARGLH 126
+G L ++ Y + G++ +++L+ + P C+I+ L W + +
Sbjct: 224 EGGLLAAES-LEDYKETFWKVGSQTLSELLHKLSSSENPPNCVIFDAFLPWVLDVGKSFG 400
Query: 127 LPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFELPGLPFSLSPSDLPSFLL-----PS 181
L + Q +V + Y+ + L++ E+ LPGLP L+P DLPSFL P
Sbjct: 401 LVGVAFFTQSCSVNSVYYHTHEKLIELPLTQSEYLLPGLP-KLAPGDLPSFLYKYGSYPG 577
Query: 182 NDSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMR-AVEEFNVIPIGPLIPSAFLDG 240
+V+ F ++ IL NS LEPE + V+ + + IGP +PS LD
Sbjct: 578 YFDIVVNQFA------NIGKADWILANSIYELEPEVVDWLVKIWPLKTIGPSVPSMLLDK 739
Query: 241 KDLNDTSFGGDIFRLSNG-CAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGH 299
+ +D +G + + C +WL+ + + SVVY SFGS LS+ Q +E+A L D
Sbjct: 740 RLKDDKEYGVSLSDPNTEFCIKWLNDKPKGSVVYASFGSMAGLSEEQTQELALGLKDSES 919
Query: 300 PFLWVIREMEEELSCREELE--EKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVS 357
FLWV+RE ++ + +E +KG +VTWC Q+ VL+H ++GCF+THCGWNST+E+L
Sbjct: 920 YFLWVVRECDQSKLPKGFVESSKKGLIVTWCPQLLVLTHEALGCFVTHCGWNSTLEALSI 1099
Query: 358 GVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAE 417
GVP++A P ++DQ+TNAK++ DVWK+GVR +E +V I+ C+ ++ EK
Sbjct: 1100GVPLIAMPLWTDQVTNAKLIADVWKMGVRA--VADEKEIVRSETIKNCIKEII-ETEKGN 1270
Query: 418 EVRRNAEKFKGLAMEAGKEGGSSEKNLMAFLA 449
E+++NA K+K LA + EGG S+KN+ F+A
Sbjct: 1271EIKKNALKWKNLAKSSVDEGGRSDKNIEEFVA 1366
>TC86702 weakly similar to GP|9794913|gb|AAF98390.1| UDP-glucose:sinapate
glucosyltransferase {Brassica napus}, partial (34%)
Length = 1366
Score = 231 bits (590), Expect = 3e-61
Identities = 154/415 (37%), Positives = 217/415 (52%), Gaps = 27/415 (6%)
Frame = +3
Query: 7 LLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPFSDG-- 64
LLI YPAQGHINP L+ AK L + GA V TT + I + +L DG
Sbjct: 84 LLISYPAQGHINPLLRLAKCLAAKGAS-VIFITTEKAGKDIRTVNNIIEKSLTSIGDGSL 260
Query: 65 ----FDDGFKLTPDTDYSL------YATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLL 114
DDG + D+ L Y LK G FV+ +I + A KPF+C+I
Sbjct: 261 TFEFLDDGL----EDDHPLRGNLLGYIEHLKLVGKPFVSQMIKNHADSNKPFSCIINNPF 428
Query: 115 LHWAAEAARGLHLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFELPGLPFS-LSPSD 173
+ W + A +P+ALLW+Q V Y YFH + LPF L ++
Sbjct: 429 VPWVCDVADEHGIPSALLWIQSTAVFSAYYNYFHKLVRFPSQTEPYIDVQLPFQVLKYNE 608
Query: 174 LPSFLLP-SNDSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVEEFNVI--PIG 230
+P FL P S F+ + EQF+ L +LV ++E LE + + + + + PIG
Sbjct: 609 IPDFLHPFSQFPFLGTLILEQFKNLSKVF--CVLVETYEELEHDFIDYISKKTIFVRPIG 782
Query: 231 PLIPSAFLDGKDLNDTSFGGDIFRLSNGC--AEWLDLQAEKSVVYVSFGSFVVLSKTQME 288
PL + + G + GD + S+ C EWL + + SVVY+SFGS V + + Q+
Sbjct: 783 PLFHNPNIKGAK----NIRGDFVK-SDDCNIIEWLSTKPKGSVVYISFGSIVYIPQEQVN 947
Query: 289 EIARALLDCGHPFLWVIREMEEELSCREE---------LEEKGKVVTWCSQVEVLSHRSV 339
EIA LLD FLWV++ EEL +E + E+GKVV W Q EVL+H SV
Sbjct: 948 EIAYGLLDSQVSFLWVLKPPSEELGLKEHNLPDGFLEGISERGKVVNWSPQEEVLAHPSV 1127
Query: 340 GCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNED 394
CF+THCGWNS+ME+L GVPM+ FP + DQ+TNAK + DV+ +G+R+ + + E+
Sbjct: 1128ACFITHCGWNSSMEALSLGVPMLTFPAWGDQVTNAKFLVDVFGVGIRLGYGMIEN 1292
>TC88485 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (39%)
Length = 1544
Score = 159 bits (401), Expect(2) = 1e-54
Identities = 90/194 (46%), Positives = 129/194 (66%), Gaps = 6/194 (3%)
Frame = +2
Query: 261 EWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIRE---MEEELSCREE 317
EWLD + + SVVYVSFG+ V + QM EI LL+ FLW + + ++ EE
Sbjct: 860 EWLDTKPKDSVVYVSFGTLVNYPQEQMNEIVYGLLNSQVSFLWSLSNPGVLPDDFL--EE 1033
Query: 318 LEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMV 377
E+GKVV W QV+VL+H SV CF+THCGWNS++E+L GVP++ FP DQ+TNAK +
Sbjct: 1034 TNERGKVVEWSPQVDVLAHPSVACFITHCGWNSSIEALSLGVPVLTFPSRGDQLTNAKFL 1213
Query: 378 EDVWKIGVRMDH--SVNEDGVVEGGEIRKC-LDAVMGNGEKAEEVRRNAEKFKGLAMEAG 434
DV+ +G++M +VN + +V E++KC L+A + GEKAE++++NA K+K A EA
Sbjct: 1214 VDVFGVGIKMGSGWAVN-NKLVTRDEVKKCLLEATI--GEKAEKLKQNANKWKKKAEEAL 1384
Query: 435 KEGGSSEKNLMAFL 448
GGSS++NL F+
Sbjct: 1385 AVGGSSDRNLDEFM 1426
Score = 72.8 bits (177), Expect(2) = 1e-54
Identities = 68/249 (27%), Positives = 108/249 (43%), Gaps = 18/249 (7%)
Frame = +1
Query: 5 RFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPFSDG 64
+ LL+ +PAQGHIN + K L + GA V TT + + + + P DG
Sbjct: 67 KLLLVSFPAQGHINHLVGLGKYLAAKGAT-VIFTTTETAGKNMRAANNIIDKLATPIGDG 243
Query: 65 ------FDDGFKLTPDTDYSLY-----ATELKRRGTEFVTDLILSAAQEGKPFTCLIYTL 113
FDDG PD D S + + E++ G ++ +I + A KPF+C+I
Sbjct: 244 TFAFEFFDDGL---PDGDRSAFRALQHSAEIEVAGRPSISQMIKNHADLNKPFSCIINNY 414
Query: 114 LLHWAAEAARGLHLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFELPGLPFSLSPS- 172
W + A ++P+ L W A V Y Y H L E P + L PS
Sbjct: 415 FFPWVCDVANEHNIPSVLSWTNSAAVFTTYYNYVHK----LTPFPTNEEPYIDVQLIPSR 582
Query: 173 -----DLPSFLLP-SNDSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVEEFNV 226
++ + P + F+ E+F++L +LV+++E LE E + + +
Sbjct: 583 VLKYNEISDLVHPFCSFPFLGKLVLEEFKDLSKVF--CVLVDTYEELEHEFIDYISK-KS 753
Query: 227 IPIGPLIPS 235
IPI + PS
Sbjct: 754 IPIRTVGPS 780
>TC86393 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyltransferase
HRA25 {Phaseolus vulgaris}, partial (47%)
Length = 1332
Score = 193 bits (490), Expect = 1e-49
Identities = 142/463 (30%), Positives = 220/463 (46%), Gaps = 15/463 (3%)
Frame = +1
Query: 1 MPHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLP----NL 56
+PH FL+I YP GHINP +Q L G + +T T +++R +N N
Sbjct: 10 IPH--FLVIPYPIPGHINPLMQLCHVLAKHGCK-ITFLNTEFSHKRTNNNNEQSQETINF 180
Query: 57 TLLPFSDGFDDGFKLTPD-TDYSLYATELKRRGTEFVTDLI--LSAAQEGKPFTCLIYTL 113
LP DG + D +D +KR + LI ++A + C+I T
Sbjct: 181 VTLP------DGLEPEDDRSDQKKVLFSIKRNMPPLLPKLIEEVNALDDENKICCIIVTF 342
Query: 114 LLHWAAEAARGLHLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFELPGLPFSLSPS- 172
+ WA E L + LLW AT L Y D ++ LSP+
Sbjct: 343 NMGWALEVGHNLGIKGVLLWTGSATSLAFCYSIPKLIDDGVIDSAGIYTKDQEIQLSPNM 522
Query: 173 ---DLPSFLLPSNDSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVEEFNVIPI 229
D + + D I + +Q + + + L N+ LE +F +PI
Sbjct: 523 PKMDTKNVPWRTFDKIIFDHLAQQMQTMKL--GHWWLCNTTYDLEHATFSISPKF--LPI 690
Query: 230 GPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEE 289
GPL+ + D N +SF ++ +WLD Q +SVVYVSFGS V+ + Q E
Sbjct: 691 GPLMEN------DSNKSSF----WQEDMTSLDWLDKQPSQSVVYVSFGSLAVMDQNQFNE 840
Query: 290 IARALLDCGHPFLWVIREMEEE----LSCREELEEKGKVVTWCSQVEVLSHRSVGCFLTH 345
+A L PFLWV+R + E L KGK+V+W Q ++L+H ++ CF++H
Sbjct: 841 LALGLDLLDKPFLWVVRPSNDNKVNYAYPDEFLGTKGKIVSWVPQKKILNHPAIACFISH 1020
Query: 346 CGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKC 405
CGWNST+E + SG+P + +P +DQ TN + DVWK+G +D +E+G+V EI+K
Sbjct: 1021CGWNSTIEGVYSGIPFLCWPFATDQFTNKSYICDVWKVGFELDK--DENGIVLKEEIKKK 1194
Query: 406 LDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
++ ++ + ++++ + K K L +E E G S KNL F+
Sbjct: 1195VEQLL----QDQDIKERSLKLKELTLENIVEDGKSSKNLQNFI 1311
>TC90534 weakly similar to GP|19911197|dbj|BAB86925. glucosyltransferase-7
{Vigna angularis}, partial (71%)
Length = 808
Score = 186 bits (473), Expect = 1e-47
Identities = 96/227 (42%), Positives = 143/227 (62%), Gaps = 3/227 (1%)
Frame = +3
Query: 225 NVIPIGPLIPSAFLDGKDLNDTSFGGDIFRL-SNGCAEWLDLQAEKSVVYVSFGSFVVLS 283
N +GP +P FLD + +D I +L S+ EWL+ + ++S VYVSFGS L+
Sbjct: 27 NFRTVGPCLPYTFLDKRVKDDEDHS--IAQLKSDESIEWLNNKPKRSAVYVSFGSMASLN 200
Query: 284 KTQMEEIARALLDCGHPFLWVIREMEEELSCR--EELEEKGKVVTWCSQVEVLSHRSVGC 341
+ Q+EE+A L DCG FLWV++ EE + E+ E G +V WC Q+EVL+H ++GC
Sbjct: 201 EEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGLIVAWCPQLEVLAHEAIGC 380
Query: 342 FLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGE 401
F+THCGWNST+E+L GVP+VA P +SDQ +AK + D+WK+G+R V+E +V
Sbjct: 381 FVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR--PLVDEKQIVRKDP 554
Query: 402 IRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
++ C+ +M EK +E+ N ++K LA A + GSS KN++ F+
Sbjct: 555 LKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFV 695
>TC91074 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (35%)
Length = 911
Score = 152 bits (383), Expect(2) = 1e-43
Identities = 81/177 (45%), Positives = 115/177 (64%), Gaps = 10/177 (5%)
Frame = +3
Query: 282 LSKTQMEEIARALLDCGHPFLWVIREMEEELSCRE---------ELEEKGKVVTWCSQVE 332
L + Q+ EIA LLD LWV++ +E +E E E+GKVV W Q E
Sbjct: 249 LPQEQVNEIAHGLLDSNVSLLWVLKPPSKESGRKEHVLPNEFLEETNERGKVVNWSPQEE 428
Query: 333 VLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVN 392
VL+H SV CF+THCGWNS+ME+L GVPM+ FP + DQ+TNAK + DV+ +G+R+ +S
Sbjct: 429 VLAHPSVACFITHCGWNSSMEALSLGVPMLTFPAWGDQVTNAKFLVDVFGVGIRLGYSHA 608
Query: 393 EDGVVEGGEIRKC-LDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
++ +V E++KC L+A + GEK EE+++NA K+K A EA GGSS++NL F+
Sbjct: 609 DNKLVTRDEVKKCLLEATI--GEKGEELKQNAIKWKKAAEEAVATGGSSDRNLDEFM 773
Score = 43.1 bits (100), Expect(2) = 1e-43
Identities = 32/89 (35%), Positives = 52/89 (57%), Gaps = 4/89 (4%)
Frame = +2
Query: 205 ILVNSFEALEPEAMRAVEEFNVI--PIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGC--A 260
+LV+SF+ LE + + + + +++ PIGPL F + K + GD + S+ C
Sbjct: 20 VLVDSFDELEHDYIDYISKKSILTRPIGPL----FNNPKIKCASDIRGDFVK-SDDCNII 184
Query: 261 EWLDLQAEKSVVYVSFGSFVVLSKTQMEE 289
EWL+ +A SVVY+SFG+ +VL T+ E
Sbjct: 185 EWLNSKANDSVVYISFGT-IVLPSTRTSE 268
>TC81622 weakly similar to PIR|T00584|T00584 indole-3-acetate
beta-glucosyltransferase homolog T27E13.12 - Arabidopsis
thaliana, partial (25%)
Length = 1735
Score = 172 bits (435), Expect = 3e-43
Identities = 127/453 (28%), Positives = 222/453 (48%), Gaps = 21/453 (4%)
Frame = +2
Query: 11 YPAQGHINPTLQFAKRLISSGAEHVTLCTTLH----TYRRISNKP------TLPNLTLLP 60
+P +GHINP L F K L S ++ + L T+ KP T+PN+ + P
Sbjct: 185 FPGRGHINPMLSFCKILTSQKPNNLLITFVLTEEWLTFIGADPKPESIRFATIPNV-IPP 361
Query: 61 FSDGFDD--GFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWA 118
+ D GF Y T+++ + + L L P ++ + L W
Sbjct: 362 EREKAGDFPGF-------YEAVMTKMEAPFEKLLDQLEL-------PVDVIVGDVELRWP 499
Query: 119 AEAARGLHLPTALLWVQPATVLDIIYYY--FHGYHDCLLSKIEFELPGLP--FSLSPSDL 174
++P A W A+ ++++ F H + K++ + +P S D+
Sbjct: 500 VNVGNRRNVPVAAFWTMSASFYSMLHHLDVFSRKHHLTVDKLDEQAENIPGISSFHIEDV 679
Query: 175 PSFLLPSNDSFIVSYFEEQFRELDVEANPTIL--VNSFEALEPEAMRAVEEFNVIPIGPL 232
+ L ND ++ ++ +AN +L V EA ++++++ F + PIGP
Sbjct: 680 QTVLC-KNDHQVLQLALGCISKVP-KANYLLLTTVQELEAETIDSLKSIFPFPIYPIGPS 853
Query: 233 IPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIAR 292
IP ++ K+ +T D + WLD Q +SV+Y+S GSF+ +S QM+EI
Sbjct: 854 IPYLDIEEKNPANTDHSQDYIK-------WLDSQPSESVLYISLGSFLSVSNAQMDEIVE 1012
Query: 293 ALLDCGHPFLWVIREMEEELSCREELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTM 352
AL + G +L+V R L +++ +KG V+ WC Q++VLSH S+G F +HCGWNST+
Sbjct: 1013ALNNSGIRYLYVARGETSRL--KDKCGDKGMVIPWCDQLKVLSHSSIGGFWSHCGWNSTL 1186
Query: 353 ESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIG--VRMDHSVNEDGVVEGGEIRKCLDAVM 410
E++ +GVP++ FP F DQ+ N+ + D WK G V + + D ++ +I + + M
Sbjct: 1187ETVFAGVPILTFPLFLDQVPNSTQIVDEWKNGWKVEIQSKLESDVILAKEDIEELVKRFM 1366
Query: 411 G-NGEKAEEVRRNAEKFKGLAMEAGKEGGSSEK 442
++ +++R A + K + +A +GGSS++
Sbjct: 1367DLENQEGKKIRDRARELKVMFRKAIGKGGSSDR 1465
>TC87122 weakly similar to GP|19911203|dbj|BAB86928. glucosyltransferase-10
{Vigna angularis}, partial (41%)
Length = 1588
Score = 171 bits (432), Expect = 7e-43
Identities = 138/482 (28%), Positives = 224/482 (45%), Gaps = 35/482 (7%)
Frame = +2
Query: 2 PHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNK------PTLPN 55
PH + + +PAQGH+NP +Q AK L G H+T ++R+ P+
Sbjct: 125 PH--VVCVPFPAQGHVNPFMQLAKILNKLGF-HITFVNNEFNHKRLIKSLGHDFLKGQPD 295
Query: 56 LTLLPFSDGFDDGFKLTPDTDYSLYATELKRRGTEFVTDLI--LSAAQEGKPFTCLIYTL 113
DG + + L ++ + +L+ L+ + E T +IY
Sbjct: 296 FRFETIPDGLPPSDENATQSIGGL-VEGCRKHCYGPLKELVENLNNSSEVPQVTSIIYDG 472
Query: 114 LLHWAAEAARGLHLPTALLWVQPATVL-------DIIYYYFHGYHD---CLLSKIEFEL- 162
L+ +A + A+ L + W A L ++I Y D ++ L
Sbjct: 473 LMGFAVDVAKDLGIAEQQFWTASACGLMGYLQFDELIKRNMLPYKDESYITDGSLDVHLD 652
Query: 163 --PGLPFSLSPSDLPSFLLPSN---DSFIVSYFEEQFRELDVEANPTILVNSFEALEPEA 217
PG+ ++ DLPSF+ + SF+ E Q + I++N+ + LE E
Sbjct: 653 WTPGMK-NIRMRDLPSFVRTTTLDEISFVGFGLEAQ----QCMKSSAIIINTVKELESEV 817
Query: 218 MRAVEEFN--VIPIGPL--IPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVY 273
+ A+ N + IGPL + + F + K+ S G +++ C +WLD SV+Y
Sbjct: 818 LDALMAINPNIYNIGPLQFLANNFPE-KENGFKSNGSSLWKNDLTCIKWLDQWEPSSVIY 994
Query: 274 VSFGSFVVLSKTQMEEIARALLDCGHPFLWVIREMEEELSCR-------EELEEKGKVVT 326
+++GS V+S+ E A L + PFLW+ R + CR +E++++G ++T
Sbjct: 995 INYGSIAVMSEKHFNEFAWGLANSKLPFLWINRPDLVKGKCRPLSQEFLDEVKDRGYIIT 1174
Query: 327 WCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVR 386
WC Q EVL+H SVG FLTHCGWNS++ES+ G PM+ +P F++Q TN + + W IG+
Sbjct: 1175WCPQSEVLAHPSVGVFLTHCGWNSSLESISEGKPMIGWPFFAEQQTNCRYISTTWGIGMD 1354
Query: 387 MDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMA 446
+ V + V E L M GEK ++ R ++K +EA K GGSS +
Sbjct: 1355IKDDVKREEVTE-------LVKEMIKGEKGKKKREKCVEWKKKVVEAAKPGGSSYNDFYR 1513
Query: 447 FL 448
L
Sbjct: 1514LL 1519
>TC89356 similar to GP|19911207|dbj|BAB86930. glucosyltransferase-12 {Vigna
angularis}, partial (58%)
Length = 1642
Score = 169 bits (427), Expect = 3e-42
Identities = 153/477 (32%), Positives = 220/477 (46%), Gaps = 35/477 (7%)
Frame = +1
Query: 2 PHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPF 61
PH L+ PAQGH+N L+ A+ L ++T T + + R L+ F
Sbjct: 61 PH--VLIFPCPAQGHVNSMLKLAELLAIQNI-YITFLNTKYIHNR-----------LIQF 198
Query: 62 SDGFDDGFKLTPDTDY---SLYATELKRRGT-EFVTDLILSAAQEGKPF----------T 107
+D + P + S + +E K G E + D+I S + GKP +
Sbjct: 199 NDDIQALLECYPKLQFKTISDFHSEEKHPGFGERIGDVITSLSLYGKPLLKDIIVSEKIS 378
Query: 108 CLIYT-----LLLHWAAE-AARGLHLPT---ALLWVQPAT--VLDIIYYYFHGYHDCLLS 156
C+I L AAE + +H T W +L+ G D +
Sbjct: 379 CIILDGIFGDLATDLAAEFGIQLIHFRTISSCCFWAYFCVPKLLECNELPIRGDED--MD 552
Query: 157 KIEFELPGLPFSLSPSDLPSFLLPSNDSFIVSYFEEQFRELDVEANPTILVNSFEALEPE 216
+I +PG+ L DLPSF + I + ++AN IL N+FE LE
Sbjct: 553 RIITNIPGMENILRCRDLPSFCRENKKDHIRLDDVALRTKQSLKANAFIL-NTFEDLEAS 729
Query: 217 AMRAVEEF--NVIPIGPLIPSAFLDGKDLNDTSF--GGDIFRLSNGCAEWLDLQAEKSVV 272
+ + + IGPL K +SF + F++ C WLD Q KSV+
Sbjct: 730 VLSQIRIHFPKLYTIGPLHHLLNTTKKSSFPSSFFSKSNFFKVDRTCMAWLDSQPLKSVI 909
Query: 273 YVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIR-EMEEELSCREELEE-----KGKVVT 326
YVSFGS + + ++ EI LL+ FLWVIR M +E ELEE KG +V
Sbjct: 910 YVSFGSTTPMKREEIIEIWHGLLNSKKQFLWVIRPNMVQEKGLLSELEEGTRKEKGLIVG 1089
Query: 327 WCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVR 386
W Q EVLSH+++G FLTH GWNST+ES+V GVPM+ +P F+DQ N++ V DVWK+G+
Sbjct: 1090WVPQEEVLSHKAIGAFLTHNGWNSTLESVVCGVPMICWPYFADQQINSRFVSDVWKLGLD 1269
Query: 387 MDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKN 443
M V + + ++ VM N + EE R+A LA ++ GGSS N
Sbjct: 1270M------KDVCDRKVVENMVNDVMVN--RKEEFVRSAMDIAKLASKSVSPGGSSYNN 1416
>TC86394 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyltransferase
HRA25 {Phaseolus vulgaris}, partial (34%)
Length = 1517
Score = 120 bits (300), Expect(2) = 7e-42
Identities = 62/174 (35%), Positives = 101/174 (57%), Gaps = 4/174 (2%)
Frame = +2
Query: 279 FVVLSKTQMEEIARALLDCGHPFLWVIREMEEE----LSCREELEEKGKVVTWCSQVEVL 334
+ V+ + Q E+A L PF+WV+R + E L KGK+V W Q ++L
Sbjct: 887 WAVMDQNQFNELALGLDLLDKPFIWVVRPSNDNKVNYAYPDEFLGTKGKIVGWAPQKKIL 1066
Query: 335 SHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNED 394
+H ++ CF++HCGWNST+E + SGVP + +P DQ N V DVWK+G+ +D +ED
Sbjct: 1067 NHPAIACFISHCGWNSTVEGVYSGVPFLCWPFHGDQFMNKSYVCDVWKVGLELDK--DED 1240
Query: 395 GVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
G++ EIR ++ ++G+ ++++ + K K L ++ E G S KNL+ F+
Sbjct: 1241 GLLPKREIRIKVEQLLGD----QDIKERSLKLKDLTLKNIVENGHSSKNLINFI 1390
Score = 68.9 bits (167), Expect(2) = 7e-42
Identities = 75/294 (25%), Positives = 117/294 (39%), Gaps = 18/294 (6%)
Frame = +3
Query: 1 MPHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYR------RISNKPTLP 54
+PH FL+I YP GH+NP +Q + L G + L T R IS K L
Sbjct: 54 VPH--FLVIPYPIAGHVNPLMQLSHLLSKHGCKITFLNTEFSNKRTNKNNISISKKDNLK 227
Query: 55 N------LTLLPFSDGFDDGFKLTPDTDYSLYATELKRRGTEFVTDLI--LSAAQEGKPF 106
N + + DG +D +D ++R + +LI ++A
Sbjct: 228 NEQSQETINFVTLPDGLEDE---DNRSDQRKVIFSIRRNMPPLLPNLIEDVNAMDAENKI 398
Query: 107 TCLIYTLLLHWAAEAARGLHLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFELPGLP 166
+C+I T + WA E L + LLW AT L Y D ++
Sbjct: 399 SCIIVTFNMGWALEVGHSLGIKGVLLWTASATSLAYCYSIPKLIDDGVMDSAGIPTTKQE 578
Query: 167 FSLSPS----DLPSFLLPSNDSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVE 222
L P+ D +F ++D + Y ++ + + + L N+ LE
Sbjct: 579 IQLFPNMPMIDTANFPWRAHDKILFDYISQEMQAM--KFGDWWLCNTTYNLEHATFSISP 752
Query: 223 EFNVIPIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSF 276
+F +PIGP F+ +D N +SF ++ C +WLD +SV YVSF
Sbjct: 753 KF--LPIGP-----FMSIED-NTSSF----WQEDATCLDWLDQYPPQSVAYVSF 878
>TC82528 weakly similar to PIR|C86356|C86356 hypothetical protein AAF87257.1
[imported] - Arabidopsis thaliana, partial (35%)
Length = 1376
Score = 164 bits (416), Expect = 5e-41
Identities = 104/310 (33%), Positives = 160/310 (51%), Gaps = 11/310 (3%)
Frame = +1
Query: 146 YFHGYHDCLLSKIEFELPGLPFSLSPSDLPSFLLPSNDSFIVSYFEEQFRELDVEANPTI 205
Y DC+ F L LP + DL F++ + FI + +QF I
Sbjct: 364 YLDNKVDCIPGMKNFRLKDLPDFIRTKDLNDFMV---EFFIEA--ADQFHRASA-----I 513
Query: 206 LVNSFEALEPEAMRAVEEF--NVIPIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWL 263
+ N++ LE + + A+ ++ IGPL PS + S ++++ C EWL
Sbjct: 514 VFNTYNELESDVLNALHSMFPSLYSIGPL-PSLLSQTPHNHLESLSSNLWKEDTKCLEWL 690
Query: 264 DLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIRE---------MEEELSC 314
+ + +SVVYV+FGS V++ Q+ E A L D PFLW+IR + E
Sbjct: 691 ESKEPESVVYVNFGSITVMTPNQLLEFAWGLADSKKPFLWIIRPDLVIGGSFILSSEFE- 867
Query: 315 REELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNA 374
E+ ++G + +WC Q +VL H S+G FLTHCGWNST ES+ +GVPM+ +P F DQ TN
Sbjct: 868 -NEISDRGLITSWCPQEQVLIHPSIGGFLTHCGWNSTTESICAGVPMLCWPFFGDQPTNC 1044
Query: 375 KMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAG 434
+ + + W+IG+ +D V D E+ K ++ + GEK +++R+ A + K A E
Sbjct: 1045RFICNEWEIGLEIDMDVKRD------EVEKLVNE-LTVGEKGKKMRQKAVELKKKAEENT 1203
Query: 435 KEGGSSEKNL 444
+ GG S NL
Sbjct: 1204RPGGRSYMNL 1233
>TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (31%)
Length = 3139
Score = 162 bits (410), Expect = 3e-40
Identities = 143/472 (30%), Positives = 226/472 (47%), Gaps = 39/472 (8%)
Frame = -1
Query: 12 PAQGHINPTLQFAKRLISSGAE-HVT-LCTTLH--TYRRISNKPTLP---NLTLLPFSDG 64
P H+ P ++FAK L+ E H+T L TL T S TLP N T+LP +
Sbjct: 2752 PGLSHLIPQVEFAKLLLQHHNESHITFLIPTLGPLTPSMQSILNTLPPNMNFTVLPQVNI 2573
Query: 65 FDDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWAAEAARG 124
D L P T L + + F+ + + S + L+ ++ A + A+
Sbjct: 2572 EDLPHNLEPSTQMKL----IVKHSIPFLHEEVKSLLSKTN-LVALVCSMFSTDAHDVAKH 2408
Query: 125 LHLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFELPGLPFSLSPSDLPSFLLPSN-- 182
+L + L+ VL + D ++ L S ++P F +P +
Sbjct: 2407 FNL-LSYLFFSSGAVLFSFFLTLPNLDDAASTQF------LGSSYEMVNVPGFSIPFHVK 2249
Query: 183 ---DSFIVSYFEEQFRE-LDVEANPT----ILVNSFEALEPEAMRAV---EEFNVIPIGP 231
D F + ++ LDV + +++N+F LE EA+R + E+ +V P+GP
Sbjct: 2248 ELPDPFNCERSSDTYKSILDVCQKSSLFDGVIINTFSNLELEAVRVLQDREKPSVFPVGP 2069
Query: 232 LIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIA 291
+I + + N+ + + C WL+ Q SV++VSFGS LS+ Q+ E+A
Sbjct: 2068 IIRN-----ESNNEANM--------SVCLRWLENQPPSSVIFVSFGSGGTLSQDQLNELA 1928
Query: 292 RALLDCGHPFLWVIREMEEELSCR------------------EELEEKGKVVT-WCSQVE 332
L GH FLWV+R + S E +EKG VVT W QVE
Sbjct: 1927 FGLELSGHKFLWVVRAPSKHSSSAYFNGQNNEPLEYLPNGFVERTKEKGLVVTSWAPQVE 1748
Query: 333 VLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVN 392
+L H S+G FL+HCGW+ST+ES+V+GVP++A+P F++Q NAK++ DV K+ VR
Sbjct: 1747 ILGHGSIGGFLSHCGWSSTLESVVNGVPLIAWPLFAEQRMNAKLLTDVLKVAVRPKVD-G 1571
Query: 393 EDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNL 444
E G+++ E+ K L +M G+++ E+R+ ++ A E GSS K L
Sbjct: 1570 ETGIIKREEVSKALKRIM-EGDESFEIRKKIKELSVSAATVLSEHGSSRKAL 1418
>CB893959 weakly similar to GP|7385017|gb| UDP-glucose:salicylic acid
glucosyltransferase {Nicotiana tabacum}, partial (28%)
Length = 791
Score = 103 bits (257), Expect(3) = 4e-40
Identities = 54/122 (44%), Positives = 79/122 (64%), Gaps = 3/122 (2%)
Frame = +3
Query: 229 IGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCA-EWLDLQAEKSVVYVSFGSFVVLSKTQM 287
IGP +PS+ LD + +D +G + + + +WL+ + ++SVVYVSFGS LS+ Q+
Sbjct: 285 IGPSVPSSHLDKRIKDDKEYGVSVSDPNTESSIKWLNEKPKRSVVYVSFGSNARLSEEQI 464
Query: 288 EEIARALLDCGHPFLWVIREMEEELSCR--EELEEKGKVVTWCSQVEVLSHRSVGCFLTH 345
EE+A L D FLWV+RE E+ + EE + G +VTWC Q+EVL+H +V CF+TH
Sbjct: 465 EELALGLNDSEKYFLWVVRESEQVKLPKGFEETSKNGLIVTWCPQLEVLTHEAVACFVTH 644
Query: 346 CG 347
CG
Sbjct: 645 CG 650
Score = 61.2 bits (147), Expect(3) = 4e-40
Identities = 25/40 (62%), Positives = 33/40 (82%)
Frame = +1
Query: 347 GWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVR 386
GWNST+E+L GVP++A P ++DQ TNAK + DVWK+GVR
Sbjct: 649 GWNSTLEALSIGVPLIAMPLWTDQATNAKFIADVWKMGVR 768
Score = 38.9 bits (89), Expect(3) = 4e-40
Identities = 30/81 (37%), Positives = 40/81 (49%), Gaps = 4/81 (4%)
Frame = +2
Query: 146 YFHGYHDCL---LSKIEFELPGLPFSLSPSDLPSFLLPSND-SFIVSYFEEQFRELDVEA 201
YFH + + +S+ E+ LPGLP L+ DLPSFL I QF ++
Sbjct: 29 YFHTHQKLIELPISQSEYLLPGLP-KLAQGDLPSFLYKYGSYPIIFDVVVNQFS--NIGK 199
Query: 202 NPTILVNSFEALEPEAMRAVE 222
IL N+F LEPE + VE
Sbjct: 200 ADWILANTFYELEPEVVGLVE 262
>TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {Rauvolfia
serpentina}, partial (34%)
Length = 1602
Score = 160 bits (406), Expect = 7e-40
Identities = 140/472 (29%), Positives = 230/472 (48%), Gaps = 39/472 (8%)
Frame = +3
Query: 12 PAQGHINPTLQFAKRLISSGAE-HVT-LCTTLH--TYRRISNKPTLP---NLTLLPFSDG 64
P H+ P ++FAK L+ + E H+T L TL T S TLP N +LP +
Sbjct: 84 PGLSHLIPLVEFAKLLLQNHNEYHITFLIPTLGPLTPSMQSILNTLPPNMNFIVLPQVNI 263
Query: 65 FDDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWAAEAARG 124
D L P T L + + F+ + + S + + L++++ A + A+
Sbjct: 264 EDLPHNLDPATQMKL----IVKHSIPFLYEEVKSLLSKTR-LVALVFSMFSTDAHDVAKH 428
Query: 125 LHLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFELPGLPFSLSPSDLPSFLLPSN-- 182
+L + L+ VL ++ + ++ L S ++P F +P +
Sbjct: 429 FNL-LSYLFFSSGAVLFSLFLTIPNLDEAASTQF------LGSSYETVNIPGFSIPLHIK 587
Query: 183 ---DSFIVSYFEEQFRE-LDVEANPT----ILVNSFEALEPEAMRAV---EEFNVIPIGP 231
D FI + ++ LDV + +++N+F LEPE +R + E+ +V P+GP
Sbjct: 588 ELPDPFICERSSDAYKSILDVCQKLSLFDGVIMNTFTDLEPEVIRVLQDREKPSVYPVGP 767
Query: 232 LIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIA 291
+I + + N+ + + C WL+ Q SV++VSFGS LS+ Q+ EIA
Sbjct: 768 MIRN-----ESNNEANM--------SMCLRWLENQQPSSVLFVSFGSGGTLSQDQLNEIA 908
Query: 292 RALLDCGHPFLWVIREMEEELSCR------------------EELEEKGKVV-TWCSQVE 332
L GH FLWV+R + S E +E G VV +W QVE
Sbjct: 909 FGLELSGHKFLWVVRAPSKNSSSAYFSGQNNDPLEYLPNGFLERTKENGLVVASWAPQVE 1088
Query: 333 VLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVN 392
+L H S+G FL+HCGW+ST+ES+V+GVP++A+P F++Q NAK++ DV K+ VR +
Sbjct: 1089ILGHGSIGGFLSHCGWSSTLESVVNGVPLIAWPLFAEQRMNAKLLTDVLKVAVRPKVD-D 1265
Query: 393 EDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNL 444
E G+++ E+ K + +M G+++ E+R+ ++ A E GSS K L
Sbjct: 1266ETGIIKQEEVAKAIKRIM-KGDESFEIRKKIKELSVGAATVLSEHGSSRKAL 1418
>TC91517 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (31%)
Length = 1027
Score = 115 bits (287), Expect(2) = 5e-39
Identities = 58/121 (47%), Positives = 86/121 (70%), Gaps = 1/121 (0%)
Frame = +1
Query: 329 SQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMD 388
S +VL+H SV CF+THCGWNS+ME++ GVPM+ FP F DQ+TNAK + DV+ G+R+
Sbjct: 523 STEQVLAHSSVACFITHCGWNSSMEAITLGVPMLTFPAFGDQLTNAKFLVDVFGAGIRLG 702
Query: 389 HSVNEDGVVEGGEIRKC-LDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAF 447
+ + +V E++KC L+A+ GEKAE +++NA K+K A +A GGSS+++L AF
Sbjct: 703 Y--GDKKLVTRDEVKKCLLEAI--TGEKAERLKQNAMKWKTAAEDAMAVGGSSDRHLDAF 870
Query: 448 L 448
+
Sbjct: 871 I 873
Score = 64.3 bits (155), Expect(2) = 5e-39
Identities = 49/170 (28%), Positives = 86/170 (49%), Gaps = 4/170 (2%)
Frame = +2
Query: 169 LSPSDLPSFLLPSNDSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVEEFNVI- 227
L +++P ++ P N I+ + D+ +LV++FE LE + + + + ++
Sbjct: 44 LKYNEIPDYIHPFNPFPILGTLTTA-QIKDMSKVFCVLVDTFEELEHDFIDYISKKSITI 220
Query: 228 -PIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCA--EWLDLQAEKSVVYVSFGSFVVLSK 284
+GPL + +G N GD + ++ EWL+ + + SVVY+SFG+ V ++
Sbjct: 221 RHVGPLFKNPKANGASNNTL---GDFTKSNDDSTIIEWLNTKPKGSVVYISFGTIVNHTQ 391
Query: 285 TQMEEIARALLDCGHPFLWVIREMEEELSCREELEEKGKVVTWCSQVEVL 334
Q+ EIA LL+ FLWV+++ EE +GKVV W Q + L
Sbjct: 392 EQVNEIAYGLLNSQVSFLWVLKQHVFPDGFLEETSGRGKVVKWSPQNKYL 541
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,314,412
Number of Sequences: 36976
Number of extensions: 233508
Number of successful extensions: 1521
Number of sequences better than 10.0: 173
Number of HSP's better than 10.0 without gapping: 1367
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1380
length of query: 457
length of database: 9,014,727
effective HSP length: 99
effective length of query: 358
effective length of database: 5,354,103
effective search space: 1916768874
effective search space used: 1916768874
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0108.10


