

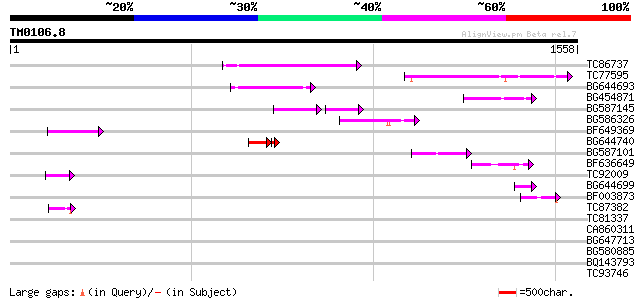
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.8
(1558 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 256 4e-68
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 176 5e-44
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 166 7e-41
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 123 6e-28
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 92 7e-28
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 104 3e-22
BF649369 87 5e-17
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 60 5e-12
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 70 5e-12
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 58 3e-08
TC92009 57 7e-08
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 54 6e-07
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 48 3e-05
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 46 1e-04
TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor... 42 0.001
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 42 0.002
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 42 0.002
BG580885 41 0.003
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 40 0.007
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 39 0.016
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 256 bits (655), Expect = 4e-68
Identities = 155/400 (38%), Positives = 231/400 (57%), Gaps = 19/400 (4%)
Frame = +1
Query: 586 VPEGMRKILEEYPEVFQEPKGLP---PRRTTDHAIQL---QEGASIPNIRPYRYPFYQKN 639
VP G +LEE+P++F K R DHAI L ++G P P+ P Y +
Sbjct: 331 VPAG---VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPP--LPWG-PLYGMS 492
Query: 640 EIEKLV-----KEMLNSGIIRHSTSPFSSPAILVKKKDGGWRFCVDYRAINKATIPDKFP 694
E LV +++L+ G I+ S S +P + V+K GG RFCVDYRA+N T D++P
Sbjct: 493 RQELLVLKKTLEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYP 672
Query: 695 IPIIDELLDEIGAAVVFSKLDLKSGYHQIRMKEEDIPKTAFRTHEGHYEYLVLPFGLTNA 754
+P+I E L + A F+KLD+ + +H++R+K+ED KTAFRT G +E++V PFGLT A
Sbjct: 673 LPLISETLRRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGA 852
Query: 755 PSTFQALMNQVLRPYLRKFVLVFFYDILIYSK-NEELHKDHLRIVLQVLKENNLVANQKK 813
P+TFQ +N+ L +L FV + D+LIY+ +++ H+ +R VL+ L + L + KK
Sbjct: 853 PATFQRYINKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKK 1032
Query: 814 CSFGQPEIIYLGHVISQA-GVAADPSKIKDMLDWPIPKEVKGLRGFLGLTGYYRRFVKNY 872
C F + Y+G +++ GV+ DP K+ + DW P VKG R FLG YY+ F+ Y
Sbjct: 1033CEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGY 1212
Query: 873 SKLAQPLNQLLKKN-SFQWTEEATQAFVKLKEVMTTVPVLVPPNFDKPFILETDASGKGL 931
S++ +PL +L +K+ F+W E AF KLK + PVL + + +ETD SG L
Sbjct: 1213SEITEPLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFAL 1392
Query: 932 GAVLMQE-----GRPVAYMSKTLSDRAQAKSVYERELMAV 966
G VL QE PVA+ S+ LS ++++EL+AV
Sbjct: 1393GGVLTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 176 bits (447), Expect = 5e-44
Identities = 140/493 (28%), Positives = 229/493 (46%), Gaps = 30/493 (6%)
Frame = +2
Query: 1085 RLLYKDRIVLPKGSTKI-------LTVLKEFHDTALGGHAGIFRTYKRISALFYWEGMKL 1137
RL ++ RI +P + +++E HD+ GH G T + +S F+W G
Sbjct: 101 RLTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQ 280
Query: 1138 DIQNYVQKCEVCQRNKYEALNPAGFLQPLPIPSQGWTDISMDFIGGLPKAMGKDT--ILV 1195
++ +V+ C+VC GFL+PLP+P++ +D+SMDFI LP G+ + + V
Sbjct: 281 TVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWV 460
Query: 1196 VVDRFTKYAHFIALSHPYNAKEIAEVFIKEVVKLHGFPTSIVSDRDRVFLSTFWSEMFKL 1255
+VDR +K + A+ A+ F+ + HG P SIVSDR ++ FW E +L
Sbjct: 461 IVDRLSKSVTLEEMD-TMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRL 637
Query: 1256 AGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGSKPKQWPKWLSWAEFWYNTNYHSAI 1315
G S++YHPQTDG TE N+ ++ LR W L + ++S+I
Sbjct: 638 TGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSI 817
Query: 1316 KTTPFKALYGRESPVIFKGNDSLTSVDEVEKWTAERNLILEELK-------SNLEKAQNR 1368
TPF +G I D+ V E E A L+++ +K + + AQ R
Sbjct: 818 GATPFFVEHGYHVDPIPTVEDTGGVVSEGE---AAAQLLVKRMKDVTGFIQAEIVAAQQR 988
Query: 1369 MRQQANKHRRDV-QYEVGDLVYLKIQPYKLKSLAKRSNQKLSPRYYGPYPIIAKINPAAY 1427
ANK R +Y+VGD V+L + YK + R ++KL ++ Y + + P
Sbjct: 989 SEASANKRRCPADRYQVGDKVWLNVSNYK----SPRPSKKLDWLHH-KYEVTRFVTPHVV 1153
Query: 1428 KLQLPEGSQVHPVFHISLLKKAVNAGVQSQPL-----PAALTEEWELKVEPEAIMDTREN 1482
+L +P V+P FH+ LL++A + + Q + P + ++ E++ E E I+ R +
Sbjct: 1154 ELNVP--GTVYPKFHVDLLRRAASDPLPGQEVVDPQPPPIVDDDGEVEWEVEEILAARWH 1327
Query: 1483 RDG---DLEVLIRWKDLPTFEDSWEDFSKLLDQFPNHQLEDKL----SLQGGRDVANPSS 1535
+ G + L++WK + +WE + + + E + G D A P
Sbjct: 1328 QVGRGRRRQALVKWKGF--VDATWEAADAIRETEALDRYEARFGPIDDHDGDPDRAAPVR 1501
Query: 1536 RPRFGN-VYARRP 1547
R R G Y RP
Sbjct: 1502 RRRGGG*CYGLRP 1540
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 166 bits (420), Expect = 7e-41
Identities = 99/243 (40%), Positives = 134/243 (54%), Gaps = 10/243 (4%)
Frame = +2
Query: 607 LPPRRTTDHAIQLQEGASIPNIRPYRYPFYQKNEIEKLV-----KEMLNSGIIRHSTSPF 661
+PP D I L +PN+ P P Y+ N ++ V K++L G I+ S P
Sbjct: 17 VPPEWKIDFGIDL-----LPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP* 181
Query: 662 SSPAILVKKKDGGWRFCVDYRAINKATIPDKFPIPIIDELLDEIGAAVVFSKLDLKSGYH 721
+ +KKKDG R +DY +N I K+P+P+IDEL D + + F K+DL+ G H
Sbjct: 182 GVVVLFLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*H 361
Query: 722 QIRMKEEDIPKTAFRTHEGHYEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDI 781
Q R+ ED+PKTAFR GHYE LV+ FG TN P F LMN+V + YL V+VF DI
Sbjct: 362 QHRVIGEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDI 541
Query: 782 LIYSKNEELHKDHLRIVLQVLKENNLVANQKKCSFGQPEII-----YLGHVISQAGVAAD 836
LIYSKNE H++HLR+ L+VLK+ L C I+ + HVIS G+ D
Sbjct: 542 LIYSKNENEHENHLRLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEGLKVD 703
Query: 837 PSK 839
+
Sbjct: 704 SKR 712
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 123 bits (308), Expect = 6e-28
Identities = 72/203 (35%), Positives = 109/203 (53%), Gaps = 1/203 (0%)
Frame = +2
Query: 1246 STFWSEMFKLAGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGSKPKQWPKWLSWAEF 1305
S FW ++FKL GT L SSAYHP +DGQ+E +N+ E YLRC+ + P +W K WAE+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1306 WYNTNYHSAIKTTPFKALYGRESPVIFKGNDSLTSVDEVEKWTAERNLILEELKSNLEKA 1365
WYNT+Y+ + TPFKALYGR+ ++ + S +++ A+R EEL S L+
Sbjct: 212 WYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQR----EELLSQLQSI 379
Query: 1366 QNRMRQQ-ANKHRRDVQYEVGDLVYLKIQPYKLKSLAKRSNQKLSPRYYGPYPIIAKINP 1424
R+ + + K R+ V + L + + + + ++
Sbjct: 380 STRLNKL*SIKLIRNAAILSSSWVSTSL*NCNLINSLR*HCGNIKSSVHP--TLVHY*QS 553
Query: 1425 AAYKLQLPEGSQVHPVFHISLLK 1447
AAYKL LP ++V P+FH+S LK
Sbjct: 554 AAYKLSLPSTAKVPPIFHVSQLK 622
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 92.0 bits (227), Expect(2) = 7e-28
Identities = 52/136 (38%), Positives = 78/136 (57%), Gaps = 3/136 (2%)
Frame = +2
Query: 725 MKEEDIPKTAFRTHEGHYEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDILIY 784
M +D+ KTAF T G Y Y V+PFGL NA ST+Q L+N++ L + V+ D+L+
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 785 SKNEELHKDHLRIVLQVLKENNLVANQKKCSFGQPEIIYLGHVISQAGVAADPSKIKDML 844
S H +HL+ + L E + N KC+FG +LG++++Q G+ +P +I +L
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 845 DWPIPK---EVKGLRG 857
D P PK EV+ L G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 52.0 bits (123), Expect(2) = 7e-28
Identities = 34/111 (30%), Positives = 53/111 (47%), Gaps = 5/111 (4%)
Frame = +3
Query: 867 RFVKNYSKLAQPLNQLLKKNS-FQWTEEATQAFVKLKEVMTTVPVLVPPNFDKPFILETD 925
RF+ + P +LL N F W E+ +AF +LK+ +TT PVL P L
Sbjct: 429 RFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIA 608
Query: 926 ASGKGLGAVLMQEGR----PVAYMSKTLSDRAQAKSVYERELMAVVLAVQK 972
S + +VL++E R P+ Y SK ++D E+ AV+ + +K
Sbjct: 609 ISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 104 bits (259), Expect = 3e-22
Identities = 76/248 (30%), Positives = 116/248 (46%), Gaps = 29/248 (11%)
Frame = +2
Query: 906 TTVPVLVPPNFDKPFILETDASGKGLGAVLMQEGRPVAYMSKTLSDRAQAKSVYERELMA 965
T+ P+LV P +++ TDAS GLG VL Q + +AY S+ L ++ E+ A
Sbjct: 8 TSAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 966 VVLAVQKWRHYLLGSQFVIHTDQRSLRFLADQRIMGEEQQKWMSKLMGYDFEIKYKPGIE 1025
VV A++ WR YL G++ IHTD +SL+++ Q + Q++WM + YD +I Y PG
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 1026 NKAADALSRK-------------------LQFSAIS------SVQCAEWADLEAEIL--- 1057
N ADALSR+ L+ + ++ ++ ADL I
Sbjct: 365 NLVADALSRRRVDVSAEREADDLDGMVRALRLNVLTKATESLGLEAVNQADLFTRIRLAQ 544
Query: 1058 -GDERYRKVLQELATQGNSAIGYQLKRGRLLYKDRIVLPKGSTKILTVLKEFHDTALGGH 1116
DE +KV Q T+ +A K G +L RI +P + ++ E H + H
Sbjct: 545 GQDENLQKVAQNDRTEYQTA-----KDGTILVNGRISVPNDRSLKEEIMSEAHKSRFSVH 709
Query: 1117 AGIFRTYK 1124
G R K
Sbjct: 710 PGAPRCIK 733
>BF649369
Length = 631
Score = 87.0 bits (214), Expect = 5e-17
Identities = 44/156 (28%), Positives = 85/156 (54%)
Frame = +3
Query: 103 VTGRRVDIPMFNGNDAYGWVTKVERFFRLSRVEEAEKIEMVMIAMEDRALGWFQWWEEQT 162
+ G++V +P+F G+D W+T+ E +F + + ++++ ++ME + WF E
Sbjct: 111 LAGKKVKLPLFEGDDPVAWITRAEIYFDVQNTPDDMRVKLSRLSMEGPTIHWFNLLMETE 290
Query: 163 LERAWEPFKQALFRRFQPALLQNPFGPLLSVKQKGSVMEYKENFELLAAPMRNADREVLK 222
+ + E K+AL R+ L+NPF L +++Q GSV E+ E FELL++ + E
Sbjct: 291 DDLSREKLKKALIARYDGRRLENPFEELSTLRQIGSVEEFVEAFELLSSQVGRLPEEQYL 470
Query: 223 GVFLNGLQEEIKAEMKLYPADDLAELMDRALLLEEK 258
G F++GL+ I+ ++ ++M A +E++
Sbjct: 471 GYFMSGLKAHIRRRVRTLNPTTRMQMMRIAKDVEDE 578
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 60.5 bits (145), Expect(2) = 5e-12
Identities = 28/63 (44%), Positives = 39/63 (61%)
Frame = -1
Query: 657 STSPFSSPAILVKKKDGGWRFCVDYRAINKATIPDKFPIPIIDELLDEIGAAVVFSKLDL 716
S SP + + V+KKDG +R C+DYR NK T +K+P+P ID L D+I F +DL
Sbjct: 256 SISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NIDL 77
Query: 717 KSG 719
+ G
Sbjct: 76 RLG 68
Score = 30.0 bits (66), Expect(2) = 5e-12
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = -3
Query: 719 GYHQIRMKEEDIPKTAFRTHEGH 741
GYHQ R+ E +IPKT T G+
Sbjct: 71 GYHQFRVNEVNIPKTTLLTWRGY 3
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 70.5 bits (171), Expect = 5e-12
Identities = 47/170 (27%), Positives = 81/170 (47%), Gaps = 4/170 (2%)
Frame = +2
Query: 1104 VLKEFHDTALGGHAGIFRTYKRIS-ALFYWEGMKLDIQNYVQKCEVCQRNKYEALN---P 1159
+L H + GH + +T +I A F+W M D +++ KC+ CQR + P
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMP 283
Query: 1160 AGFLQPLPIPSQGWTDISMDFIGGLPKAMGKDTILVVVDRFTKYAHFIALSHPYNAKEIA 1219
F+ + + W +DF+G P + ILV VD +K+ IA S +A +
Sbjct: 284 QNFILEVEV-FDVW---GIDFMGPFPSSYNNKYILVAVDYVSKWVEAIA-SPTNDATVVV 448
Query: 1220 EVFIKEVVKLHGFPTSIVSDRDRVFLSTFWSEMFKLAGTKLKFSSAYHPQ 1269
++F + G P ++SD F++ + ++ K G + K ++AYHPQ
Sbjct: 449 KMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 57.8 bits (138), Expect = 3e-08
Identities = 51/182 (28%), Positives = 78/182 (42%), Gaps = 13/182 (7%)
Frame = +1
Query: 1270 TDGQTEVVNRCVETYLRCVTGSKPKQWPKWLSWAEFWYNTNYHSAIKTTPFKALYGRESP 1329
+D Q ++N +ET+L T + +L+WAE YNTN+H TPFK +Y
Sbjct: 7 SDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY----- 171
Query: 1330 VIFKGNDSLTSVDEVEKWTAERNLIL--EELKSNLEKAQNRMRQQANKHRRDVQYEVG-- 1385
V ++K+ R+LI E L + +A + YE
Sbjct: 172 ----------VVAHLQKFVVARDLIYRNEGLHKSST*TSFGRGTRAYEALSRPAYETC*H 321
Query: 1386 -----DLVYLKIQPYKLKSLAKRSNQKLSPRY----YGPYPIIAKINPAAYKLQLPEGSQ 1436
+VY + + Y+ + L P+Y YGPY +I +I A+KL LPE Q
Sbjct: 322 PRRPLSIVYTRDRTYEWQVL---------PKYVA*CYGPYQVIKQIGSVAFKL*LPEQHQ 474
Query: 1437 VH 1438
+H
Sbjct: 475 IH 480
>TC92009
Length = 974
Score = 56.6 bits (135), Expect = 7e-08
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Frame = +2
Query: 98 SHLRAVTGRRVDIPMFNGNDAYGWVTKVERFFRLSRVEEAEKIEMVMIAMED-RALGWFQ 156
S + + + + F G D GW+T+ E FF + + +K++ ++MED +AL WF
Sbjct: 653 SEMDGIWTLKYKLSQFTGTDPAGWITRAEMFFADNEIHSCDKLQWAFMSMEDEKALLWFY 832
Query: 157 WWEEQTLERAWEPFKQALFRRF 178
W ++ + W+ F A+ R F
Sbjct: 833 SWCQENPDADWKSFSMAMIREF 898
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 53.5 bits (127), Expect = 6e-07
Identities = 28/62 (45%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Frame = +2
Query: 1388 VYLKIQPYKLKSLAKRSNQKLSPRYYGPYPIIAKINPAAYKLQLPEG-SQVHPVFHISLL 1446
V LK+ P + KLS RY GP+ +I +I AY+L LP G S VHPVFH+S+
Sbjct: 8 VLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVSMF 187
Query: 1447 KK 1448
K+
Sbjct: 188 KR 193
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 48.1 bits (113), Expect = 3e-05
Identities = 34/122 (27%), Positives = 62/122 (49%), Gaps = 12/122 (9%)
Frame = +2
Query: 1405 NQKLSPRYYGPYPIIAKINPAAYKLQLPEG-SQVHPVFHISLLKKAVNAGVQSQPLPAAL 1463
++KL+ R+ GPY I ++ AY++ LP +H VFH+S L+K V P P+ +
Sbjct: 23 SKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYV-------PDPSHV 181
Query: 1464 TEEWELKVEPEAIMDTRENRDGDLEV-LIRWKDLPTFED----------SWEDFSKLLDQ 1512
+ +++V ++T R D +V +R K++P +WE SK+++
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 1513 FP 1514
+P
Sbjct: 362 YP 367
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 45.8 bits (107), Expect = 1e-04
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 13/87 (14%)
Frame = +2
Query: 107 RVDIPMFNGN----DAYGWVTKVERFFRLSRVEEAEKIEMVMIAMEDRALGWFQWWEEQT 162
+VDIP F GN D W+ +ER F V E +K+++V ++ AL WWE
Sbjct: 617 KVDIPDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHAL---IWWENLK 787
Query: 163 LER---------AWEPFKQALFRRFQP 180
R W+ +Q L R++ P
Sbjct: 788 RRRKREGKSKIKTWDKMRQKLTRKYLP 868
>TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor small
subunit {Arabidopsis thaliana}, partial (81%)
Length = 1190
Score = 42.4 bits (98), Expect = 0.001
Identities = 27/61 (44%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Frame = +2
Query: 32 ELRRLFLSRDRRRTRGRSNTPRHRRSSREHNSVSTA-RTYD--GSRTGSRTGSRTASRSR 88
ELRR +S R R+R RSN+PR RR SR R YD G R+ R R R R
Sbjct: 617 ELRRKLVSSQRSRSRSRSNSPRRRRRSRSRERERPRDRDYDSRGRRSSDRDRVRDGDRDR 796
Query: 89 E 89
E
Sbjct: 797 E 799
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 42.0 bits (97), Expect = 0.002
Identities = 22/58 (37%), Positives = 35/58 (59%), Gaps = 5/58 (8%)
Frame = +1
Query: 930 GLGAVLMQEGR-----PVAYMSKTLSDRAQAKSVYERELMAVVLAVQKWRHYLLGSQF 982
G+GA L Q+ P+AY S+ L+ + +V ERE +A + A++ +RHYL G +F
Sbjct: 13 GIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKF 186
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 42.0 bits (97), Expect = 0.002
Identities = 35/131 (26%), Positives = 62/131 (46%), Gaps = 11/131 (8%)
Frame = +2
Query: 68 RTYDGSRTGSRTGSRTASRSREHHEHYHHRSHLRAVTGRRVDIPMFNG----NDAYGWVT 123
R D +GS + S +SRS HR R ++ +VDIP F G ++ W+
Sbjct: 245 RRNDDDGSGSDSSSSRSSRS--------HRRQTR-MSKIKVDIPDF*GKLQPDEFVDWLQ 397
Query: 124 KVERFFRLSRVEEAEKIEMVMIAMEDRALGWF------QWWEEQTLERAWEPFKQALFRR 177
+ER F+ V E +K+++V ++ A W+ + E ++ + W+ +Q L R+
Sbjct: 398 TIERVFKYKEVAEEQKVKIVAAKLKKHASIWWKNLKRKRNCEGKSKIKTWDKMRQKLTRK 577
Query: 178 F-QPALLQNPF 187
+ P Q+ F
Sbjct: 578 YLHPHYYQDNF 610
>BG580885
Length = 609
Score = 41.2 bits (95), Expect = 0.003
Identities = 32/133 (24%), Positives = 62/133 (46%), Gaps = 10/133 (7%)
Frame = +3
Query: 110 IPMFNGNDAYGWVTKVERFFRLSRVEEAEKIEMVM----IAMEDRALGWFQW-WEEQTLE 164
+P+F G +T + RF ++ R A +EM + +E+ + W+ E +
Sbjct: 45 LPIFRGTPNESPITHLSRFNKVCRANNASSVEMQKKIFPVTLEEESALWYDLNIEPYYIS 224
Query: 165 RAWEPFKQALFRRF-QPALLQNPFGPLLSVKQ--KGSVMEY--KENFELLAAPMRNADRE 219
+W+ K + + + + ++ L+ + Q K V Y + + L P + +
Sbjct: 225 LSWDEIKLSFLQAYYEIEPVEELRSELMGIHQGEKERVRSYFLRLQWILKRWPEHGLEDD 404
Query: 220 VLKGVFLNGLQEE 232
V+KGVF+NGL+EE
Sbjct: 405 VIKGVFVNGLREE 443
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 40.0 bits (92), Expect = 0.007
Identities = 31/107 (28%), Positives = 41/107 (37%)
Frame = -3
Query: 263 RGGKPKEEDKRGWKEKGGVGGRYYSSTGDSMGRIANSYVNFQSKGGTGNQDNEGKSLQNK 322
RGG KE+ KRGW+ KG GD + GG G G +
Sbjct: 351 RGGGKKEKKKRGWEGKGEG*NNIGEGRGDGK----------RKWGGGGGGRRRGGEGMGR 202
Query: 323 GGTGNQDTEGKQPEKKWNGGQRLTQTELQERSRKGLCFKCGDKWGKE 369
GG G + K K+W G R+ + E +G C +WG E
Sbjct: 201 GGGG*MIFKEKGKGKRWGG*DRVGERE----GGRGRGDGCYGEWGGE 73
Score = 32.0 bits (71), Expect = 1.9
Identities = 30/106 (28%), Positives = 46/106 (43%), Gaps = 13/106 (12%)
Frame = -3
Query: 264 GGKPKEEDKRGWKEKGGVGGRYYSSTGDSMGRIANSYVNFQSKGGTGNQDNEGKSL---- 319
GG+ K++ KRG G GGR GD G + + K N++ G+
Sbjct: 681 GGRLKQKKKRG-----GGGGRGGGGRGD--GIVVREVRVKRRKMDGANEERWGRERGRVN 523
Query: 320 ----QNKGGTGNQDTEGKQPE-----KKWNGGQRLTQTELQERSRK 356
+ +GGT + EGK+ E K+ GG+R + +E S K
Sbjct: 522 GNRGKGRGGTSERKEEGKEKEERGKRKEGKGGERKGEEGEEEGSGK 385
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 38.9 bits (89), Expect = 0.016
Identities = 23/67 (34%), Positives = 34/67 (50%), Gaps = 14/67 (20%)
Frame = +2
Query: 1155 EALNPAGFLQPL-------------PIPSQGWTDISMDF-IGGLPKAMGKDTILVVVDRF 1200
E P GFL + P+P W D+++DF +G L KD+ +VV D+F
Sbjct: 398 EEFKPLGFLADMEPFKDKTKLCLLRPVPKPPWEDVTIDFSLGLL*TQQLKDSKMVVGDKF 577
Query: 1201 TKYAHFI 1207
++ AHFI
Sbjct: 578 SRMAHFI 598
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,672,488
Number of Sequences: 36976
Number of extensions: 613754
Number of successful extensions: 4473
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 4036
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4352
length of query: 1558
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1449
effective length of database: 4,984,343
effective search space: 7222313007
effective search space used: 7222313007
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0106.8


