

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.13
(445 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
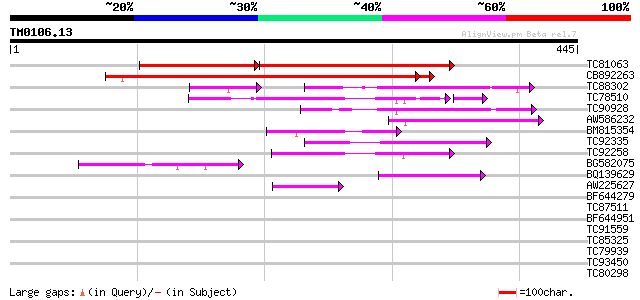
Score E
Sequences producing significant alignments: (bits) Value
TC81063 similar to GP|13877621|gb|AAK43888.1 Unknown protein {Ar... 299 e-129
CB892263 similar to GP|13877621|gb Unknown protein {Arabidopsis ... 411 e-115
TC88302 similar to GP|8953714|dbj|BAA98077.1 contains similarity... 59 3e-11
TC78510 similar to GP|6648201|gb|AAF21199.1| putative GTPase act... 55 6e-11
TC90928 homologue to GP|21593435|gb|AAM65402.1 putative plant ad... 64 2e-10
AW586232 similar to PIR|T05242|T05 hypothetical protein F18A5.12... 56 2e-08
BM815354 similar to PIR|T05242|T05 hypothetical protein F18A5.12... 54 1e-07
TC92335 similar to GP|14517422|gb|AAK62601.1 AT5g53570/MNC6_11 {... 51 1e-06
TC92258 similar to GP|9758258|dbj|BAB08757.1 contains similarity... 50 2e-06
BG582075 similar to PIR|T05242|T052 hypothetical protein F18A5.1... 49 4e-06
BQ139629 similar to GP|15408841|dbj hypothetical protein~similar... 47 1e-05
AW225627 similar to GP|18377650|gb unknown protein {Arabidopsis ... 45 4e-05
BF644279 similar to PIR|T47641|T47 hypothetical protein T15C9.20... 39 0.003
TC87511 similar to GP|17381090|gb|AAL36357.1 unknown protein {Ar... 32 0.64
BF644951 similar to GP|16550928|gb calcineurin-like protein {Euc... 30 1.4
TC91559 similar to GP|10178129|dbj|BAB11541. gene_id:MOP10.6~unk... 28 5.5
TC85325 28 7.1
TC79939 similar to GP|9758423|dbj|BAB08965.1 gene_id:MHF15.5~sim... 28 7.1
TC93450 similar to GP|8777354|dbj|BAA96944.1 microtubule-associa... 28 9.3
TC80298 weakly similar to GP|14517422|gb|AAK62601.1 AT5g53570/MN... 28 9.3
>TC81063 similar to GP|13877621|gb|AAK43888.1 Unknown protein {Arabidopsis
thaliana}, partial (53%)
Length = 749
Score = 299 bits (765), Expect(2) = e-129
Identities = 144/153 (94%), Positives = 151/153 (98%)
Frame = +2
Query: 197 ERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLV 256
ERS+DEI+MLRQIAVDCPRTVPDV+FFQQ QVQKSLERILYAWAIRHPASGYVQGINDLV
Sbjct: 290 ERSDDEISMLRQIAVDCPRTVPDVAFFQQPQVQKSLERILYAWAIRHPASGYVQGINDLV 469
Query: 257 TPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQR 316
TPFFVVFLSEYLEGSI+NW+MSDLSSD+ISNVEADCYWCLSKLLDGMQDHYTFAQPGIQR
Sbjct: 470 TPFFVVFLSEYLEGSINNWTMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQR 649
Query: 317 LVFKLKELVRRIDDPVSTHMENQGLEFLQFAFR 349
LVFKLKELVRRIDDPVS+HMENQGLEFL FAFR
Sbjct: 650 LVFKLKELVRRIDDPVSSHMENQGLEFLHFAFR 748
Score = 181 bits (459), Expect(2) = e-129
Identities = 86/94 (91%), Positives = 91/94 (96%)
Frame = +1
Query: 103 ENPSEDVRKSSMGARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLL 162
ENP+E++RKSS+G R TDSARVMKFTKVLSGT+VILDKLRELAWSGVPDYMRPTVWRLLL
Sbjct: 7 ENPTEEIRKSSIGGRTTDSARVMKFTKVLSGTMVILDKLRELAWSGVPDYMRPTVWRLLL 186
Query: 163 GYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDT 196
GYAP NSDRREGVLRRKRLEYLDCVSQYYDIPDT
Sbjct: 187 GYAPTNSDRREGVLRRKRLEYLDCVSQYYDIPDT 288
>CB892263 similar to GP|13877621|gb Unknown protein {Arabidopsis thaliana},
partial (47%)
Length = 804
Score = 411 bits (1057), Expect(2) = e-115
Identities = 205/250 (82%), Positives = 222/250 (88%), Gaps = 3/250 (1%)
Frame = +2
Query: 76 SETVEVEVHSSS---GIPGENKLKISTSHVENPSEDVRKSSMGARATDSARVMKFTKVLS 132
S VE EV S+S +KLK STS EN SED+RK ++GARATDSARV KF KVLS
Sbjct: 8 SGAVEEEVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSARVTKFNKVLS 187
Query: 133 GTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYD 192
GTVVILD LRELAWSGVPDYMRP VWRLLLGY PPNSDR+EGVL RKR EYLDC+SQYYD
Sbjct: 188 GTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYD 367
Query: 193 IPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGI 252
IPD+ERS+DE+NMLRQIAVDCPRTVPDV+FFQQQQVQKSLERILYAWAIRHPASGYVQGI
Sbjct: 368 IPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGI 547
Query: 253 NDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQP 312
NDLVTPF VVF+SE+LEG ID+WSMSDLSSD+ISNVEADCYWCLSKLLDGMQ+HYTFAQP
Sbjct: 548 NDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQEHYTFAQP 727
Query: 313 GIQRLVFKLK 322
GIQRL F L+
Sbjct: 728 GIQRLGFNLR 757
Score = 23.5 bits (49), Expect(2) = e-115
Identities = 9/15 (60%), Positives = 13/15 (86%)
Frame = +1
Query: 319 FKLKELVRRIDDPVS 333
F +ELVR+ID+P+S
Sbjct: 745 F*FEELVRKIDEPIS 789
>TC88302 similar to GP|8953714|dbj|BAA98077.1 contains similarity to GTPase
activating protein~gene_id:F6N7.7 {Arabidopsis
thaliana}, complete
Length = 1198
Score = 58.9 bits (141), Expect(2) = 3e-11
Identities = 42/183 (22%), Positives = 77/183 (41%), Gaps = 2/183 (1%)
Frame = +1
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEAD 291
L IL ++ + GY QG++DL++P V M D E++
Sbjct: 406 LRDILLTYSFYNFDLGYCQGMSDLLSPILFV--------------MED---------ESE 516
Query: 292 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWF 351
+WC L++ + ++ Q G+ +F L +LV +D P+ + + + F FRW
Sbjct: 517 AFWCFVSLMERLGPNFNRDQNGMHSQLFALSKLVELLDSPLHNYFKQRDCLNYFFCFRWI 696
Query: 352 NCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKL--QKLDFQDLVMFL 409
RE + RLW+ + +Y+ + L K+ +++DF L+ F+
Sbjct: 697 LIQFKREFEYEKTMRLWEVLWTHYPS-EHLHLYVCVAVLKRCRGKIIGEEMDFDSLLKFI 873
Query: 410 QHL 412
L
Sbjct: 874 NEL 882
Score = 26.9 bits (58), Expect(2) = 3e-11
Identities = 19/58 (32%), Positives = 27/58 (45%), Gaps = 2/58 (3%)
Frame = +3
Query: 142 RELAWSGVPDYMRPTVWRLLLGYAPPNSD--RREGVLRRKRLEYLDCVSQYYDIPDTE 197
RE + + R VW LLLGY P +S RE + K+ EY +Q+ I +
Sbjct: 117 REYFTADLTTNYRNEVWGLLLGYYPYDSTYAEREFLKSVKKSEYETIKNQWQSISSAQ 290
>TC78510 similar to GP|6648201|gb|AAF21199.1| putative GTPase activator
protein {Arabidopsis thaliana}, partial (92%)
Length = 2077
Score = 55.1 bits (131), Expect(2) = 6e-11
Identities = 55/211 (26%), Positives = 91/211 (43%), Gaps = 5/211 (2%)
Frame = +2
Query: 141 LRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTERSE 200
L++L G+P +RP VW L G A S + YYD T+ E
Sbjct: 917 LKKLIRKGIPPVLRPKVWFSLSGAAKKKSTVPDS---------------YYD-DLTKAVE 1048
Query: 201 DEIN-MLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPF 259
++ RQI D PRT P ++ + +L R+L A++ R GY QG+N +
Sbjct: 1049 GKVTPATRQIDHDLPRTFPGHAWLDTPEGHAALRRVLVAYSFRDSDVGYCQGLNYVAALL 1228
Query: 260 FVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDG--MQDHYT--FAQPGIQ 315
+V E D +W L+ LL+ + D YT ++ ++
Sbjct: 1229 LLVM-----------------------KTEEDAFWMLAVLLENVLVSDCYTNNLSRCHVE 1339
Query: 316 RLVFKLKELVRRIDDPVSTHMENQGLEFLQF 346
+ VF K+L+ + ++TH+++ LEF +F
Sbjct: 1340 QRVF--KDLLTKKCPRIATHLDS--LEFDRF 1420
Score = 29.6 bits (65), Expect(2) = 6e-11
Identities = 10/27 (37%), Positives = 14/27 (51%)
Frame = +1
Query: 349 RWFNCLLIREIPFNMVTRLWDTYLAEG 375
+WF CL + +P R+WD EG
Sbjct: 1435 KWFLCLFSKSLPSETTLRVWDVIFYEG 1515
>TC90928 homologue to GP|21593435|gb|AAM65402.1 putative plant adhesion
molecule {Arabidopsis thaliana}, partial (66%)
Length = 1058
Score = 63.5 bits (153), Expect = 2e-10
Identities = 49/190 (25%), Positives = 81/190 (41%), Gaps = 5/190 (2%)
Frame = +2
Query: 229 QKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNV 288
Q+SL +L A+++ GYVQG+ F L Y+
Sbjct: 131 QRSLYNVLKAYSVFDREVGYVQGMG-----FLAGLLLLYMS------------------- 238
Query: 289 EADCYWCLSKLLDG-----MQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEF 343
E D +W L LL G M+ Y P +Q+ +F+ + LVR + H + +
Sbjct: 239 EEDAFWLLVALLKGAVHAPMEGLYLAGLPLVQQYLFQFERLVREHLPKLGEHFTQEMINP 418
Query: 344 LQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQ 403
+A +WF + PF++ R+WD +L EG + + + + L D L KL F+
Sbjct: 419 SMYASQWFITVFSYSFPFHLALRIWDVFLYEGVKI---VFKVGLALLKYCHDDLVKLPFE 589
Query: 404 DLVMFLQHLP 413
L+ L++ P
Sbjct: 590 KLIHALKNFP 619
>AW586232 similar to PIR|T05242|T05 hypothetical protein F18A5.120 -
Arabidopsis thaliana, partial (30%)
Length = 529
Score = 56.2 bits (134), Expect = 2e-08
Identities = 36/127 (28%), Positives = 65/127 (50%), Gaps = 5/127 (3%)
Frame = +1
Query: 298 KLLDGMQDHYTF----AQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQF-AFRWFN 352
+LL G++D++ + GI+ + KL +L+R+ D+ + H+E QF AFRW
Sbjct: 4 ELLSGLRDNFVQQLDNSVVGIRSTITKLSQLLRKHDEELWRHLEITSKINPQFYAFRWIT 183
Query: 353 CLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHL 412
LL +E F +WDT L + + + L+ + + L+ +L DF + LQ+
Sbjct: 184 LLLTQEFNFADSLHIWDTLLGDPEGPQETLLRVCCAMLILIRKRLLAGDFTSNLKLLQNY 363
Query: 413 PTQDWTH 419
P+ + +H
Sbjct: 364 PSTNISH 384
>BM815354 similar to PIR|T05242|T05 hypothetical protein F18A5.120 -
Arabidopsis thaliana, partial (31%)
Length = 494
Score = 53.9 bits (128), Expect = 1e-07
Identities = 33/111 (29%), Positives = 57/111 (50%), Gaps = 5/111 (4%)
Frame = +3
Query: 202 EINMLRQIAVDCPRTVPDVSFF-----QQQQVQKSLERILYAWAIRHPASGYVQGINDLV 256
+ +++ QI D RT PD+ FF + Q++L+ IL +A +P YVQG+N+++
Sbjct: 177 DTDIIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVL 356
Query: 257 TPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHY 307
P F VF +D + + EAD ++C +LL G +D++
Sbjct: 357 APLFYVF-------------KNDPDEENAAFSEADTFFCFVELLSGFRDNF 470
>TC92335 similar to GP|14517422|gb|AAK62601.1 AT5g53570/MNC6_11 {Arabidopsis
thaliana}, partial (37%)
Length = 1077
Score = 50.8 bits (120), Expect = 1e-06
Identities = 33/147 (22%), Positives = 62/147 (41%)
Frame = +1
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEAD 291
L IL A+A+ P GY QG++DL++P V ++ +
Sbjct: 13 LVAILEAYALYDPEIGYCQGMSDLLSPIICVVSEDH-----------------------E 123
Query: 292 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWF 351
+WC + + ++ + GI+R + + ++++ D + H+E E F +R
Sbjct: 124 AFWCFVGFMKKARQNFRLDEVGIRRQLDIVAKIIKFKDSHLFRHLEKLQAEDCFFVYRMV 303
Query: 352 NCLLIREIPFNMVTRLWDTYLAEGDAL 378
L RE+ F LW+ A+ A+
Sbjct: 304 VVLFRRELTFEQTLCLWEVMWADQAAI 384
>TC92258 similar to GP|9758258|dbj|BAB08757.1 contains similarity to GTPase
activating protein~gene_id:MBG8.4 {Arabidopsis
thaliana}, partial (38%)
Length = 727
Score = 50.1 bits (118), Expect = 2e-06
Identities = 37/146 (25%), Positives = 65/146 (44%), Gaps = 2/146 (1%)
Frame = +1
Query: 206 LRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLS 265
L QI +D RT + F+++Q+ L IL +A GY QG++DL +P ++
Sbjct: 349 LHQIGLDVVRTDRTLVFYEKQENLSKLWDILAVYAWIDKEVGYGQGMSDLCSPMIILL-- 522
Query: 266 EYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHY--TFAQPGIQRLVFKLKE 323
+ EAD +WC +L+ ++ ++ T G++ + L
Sbjct: 523 ---------------------DDEADAFWCFERLMRRLRGNFRCTGRTLGVEAQLSNLAS 639
Query: 324 LVRRIDDPVSTHMENQGLEFLQFAFR 349
+ + ID + H+E+ G FAFR
Sbjct: 640 ITQVIDPKLHKHIEHIGGGDYVFAFR 717
>BG582075 similar to PIR|T05242|T052 hypothetical protein F18A5.120 -
Arabidopsis thaliana, partial (21%)
Length = 784
Score = 48.9 bits (115), Expect = 4e-06
Identities = 38/138 (27%), Positives = 62/138 (44%), Gaps = 9/138 (6%)
Frame = +1
Query: 55 LSSPTYTRSNSYNDAGTSDHASETVEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSM 114
L+SP ++ N + + SE + P I+TS + S SS
Sbjct: 85 LNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSS-----SSA 249
Query: 115 GARATDSARVMKFTKV-------LSGTVVILDKLRELAWSGVPDY--MRPTVWRLLLGYA 165
+TD + + + LS V+ + +LR++A G+PD +R T+W+LLLGY
Sbjct: 250 STSSTDDMSISRLAQSQSQLLAELSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYL 429
Query: 166 PPNSDRREGVLRRKRLEY 183
PP+ L +KR +Y
Sbjct: 430 PPDRSLWSSELAKKRSQY 483
>BQ139629 similar to GP|15408841|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 2 At2g43490, partial
(15%)
Length = 666
Score = 47.4 bits (111), Expect = 1e-05
Identities = 25/85 (29%), Positives = 40/85 (46%), Gaps = 1/85 (1%)
Frame = +1
Query: 290 ADCYWCLSKLLDGMQDHYTFAQPG-IQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAF 348
AD +WC LL M++++ P + + + L ++ D + H+ G E L FAF
Sbjct: 34 ADAFWCFEMLLRRMRENFKMEGPTRVMKQLRALWHILELSDKELFAHLSKIGAESLHFAF 213
Query: 349 RWFNCLLIREIPFNMVTRLWDTYLA 373
R L RE+ FN +W+ A
Sbjct: 214 RMLLVLFRRELSFNEALSMWEVRAA 288
>AW225627 similar to GP|18377650|gb unknown protein {Arabidopsis thaliana},
partial (16%)
Length = 455
Score = 45.4 bits (106), Expect = 4e-05
Identities = 23/57 (40%), Positives = 32/57 (55%), Gaps = 1/57 (1%)
Frame = +3
Query: 207 RQIAVDCPRTVPDV-SFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVV 262
R + D R P+ S+FQ Q L RIL W ++HP GY QG+++L+ PF V
Sbjct: 42 RLVDQDLSRLYPEHGSYFQTPGCQGMLRRILLLWCLKHPDCGYRQGMHELLAPFLYV 212
>BF644279 similar to PIR|T47641|T47 hypothetical protein T15C9.20 -
Arabidopsis thaliana, partial (20%)
Length = 681
Score = 39.3 bits (90), Expect = 0.003
Identities = 30/114 (26%), Positives = 51/114 (44%), Gaps = 5/114 (4%)
Frame = +3
Query: 314 IQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLA 373
+ +LVF+ ELVR ++ H+ G++ WF + + +P+ V R+WD L
Sbjct: 24 VDQLVFE--ELVRERFPKLANHLXYLGVQVAWVTGPWFLSIFVNMLPWESVLRVWDVLLF 197
Query: 374 EGDALPDF-----LVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQEL 422
EG+ + F L+ ++ L+T D D V LQ L + +L
Sbjct: 198 EGNRVMLFXTAVALMELYGPALVTTKDA------GDAVTLLQSLAGSTFDSSQL 341
>TC87511 similar to GP|17381090|gb|AAL36357.1 unknown protein {Arabidopsis
thaliana}, partial (44%)
Length = 891
Score = 31.6 bits (70), Expect = 0.64
Identities = 19/68 (27%), Positives = 34/68 (49%), Gaps = 10/68 (14%)
Frame = +2
Query: 349 RWFN----CLLIREIPFNMVT-----RLWDTYLAEGDAL-PDFLVYIFASFLLTWSDKLQ 398
+WF CLL+ E PFN +T ++ +L +G P F++YI + L ++
Sbjct: 623 KWFTFGKVCLLVLEDPFNEMTWFLGLSIFYFFLDQGFGFPPPFVIYIIVAVLCSYGFSFS 802
Query: 399 KLDFQDLV 406
K++ +V
Sbjct: 803 KMELNTMV 826
>BF644951 similar to GP|16550928|gb calcineurin-like protein {Eucalyptus
grandis}, complete
Length = 677
Score = 30.4 bits (67), Expect = 1.4
Identities = 16/49 (32%), Positives = 29/49 (58%)
Frame = +1
Query: 377 ALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMV 425
++P+F++ + LL D L +F+D V FL T+ THQ++E++
Sbjct: 223 SIPEFVMNPLSQRLLKMVDGL---NFKDFVAFLSAFSTKAGTHQKVELI 360
>TC91559 similar to GP|10178129|dbj|BAB11541. gene_id:MOP10.6~unknown
protein {Arabidopsis thaliana}, partial (24%)
Length = 720
Score = 28.5 bits (62), Expect = 5.5
Identities = 13/37 (35%), Positives = 20/37 (53%)
Frame = +2
Query: 261 VVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLS 297
V LS ++E + +S L S + SN+ A C+W S
Sbjct: 449 VQLLSSFIEAGLTTEILSFLCSSQASNIVALCHWMAS 559
>TC85325
Length = 3584
Score = 28.1 bits (61), Expect = 7.1
Identities = 13/42 (30%), Positives = 25/42 (58%)
Frame = +3
Query: 350 WFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLL 391
+ CL I E + +T++WD +L D + + +++F+ FLL
Sbjct: 2451 YLKCLEILESRYIKITKIWDLFL*HADWIVN--LFLFSIFLL 2570
>TC79939 similar to GP|9758423|dbj|BAB08965.1 gene_id:MHF15.5~similar to
unknown protein~sp|P73920 {Arabidopsis thaliana},
partial (57%)
Length = 981
Score = 28.1 bits (61), Expect = 7.1
Identities = 13/28 (46%), Positives = 19/28 (67%)
Frame = -3
Query: 356 IREIPFNMVTRLWDTYLAEGDALPDFLV 383
+RE +NM T +WDT L++G P F+V
Sbjct: 619 MRESSYNM*TTMWDTRLSQG---PSFVV 545
>TC93450 similar to GP|8777354|dbj|BAA96944.1 microtubule-associated
protein-like {Arabidopsis thaliana}, partial (22%)
Length = 806
Score = 27.7 bits (60), Expect = 9.3
Identities = 15/66 (22%), Positives = 27/66 (40%)
Frame = +3
Query: 312 PGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTY 371
P + L L+ D + +H+ + +E F RW L RE + + +WD
Sbjct: 84 PPVIEASMALYHLLSLADSSLHSHLLDLEVEPQYFYLRWLRVLFGREFSLDKLLVIWDEI 263
Query: 372 LAEGDA 377
A ++
Sbjct: 264 FASDNS 281
>TC80298 weakly similar to GP|14517422|gb|AAK62601.1 AT5g53570/MNC6_11
{Arabidopsis thaliana}, partial (15%)
Length = 981
Score = 27.7 bits (60), Expect = 9.3
Identities = 19/74 (25%), Positives = 33/74 (43%), Gaps = 2/74 (2%)
Frame = +2
Query: 141 LRELAWSGVPDYMRPTVWRLLLGYAPPNS--DRREGVLRRKRLEYLDCVSQYYDIPDTER 198
L+ + GV +R VW LLG NS D R+ V + R +Y + Q +
Sbjct: 449 LKRVRNGGVHPRIRAEVWPFLLGVYDFNSTKDERDAVKTQNRKQYEELRRQCTKLIKQSN 628
Query: 199 SEDEINMLRQIAVD 212
++N + +I+ +
Sbjct: 629 ENSKLNEIGEISYE 670
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,902,829
Number of Sequences: 36976
Number of extensions: 186906
Number of successful extensions: 1000
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 959
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 995
length of query: 445
length of database: 9,014,727
effective HSP length: 99
effective length of query: 346
effective length of database: 5,354,103
effective search space: 1852519638
effective search space used: 1852519638
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0106.13


