

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0105.15
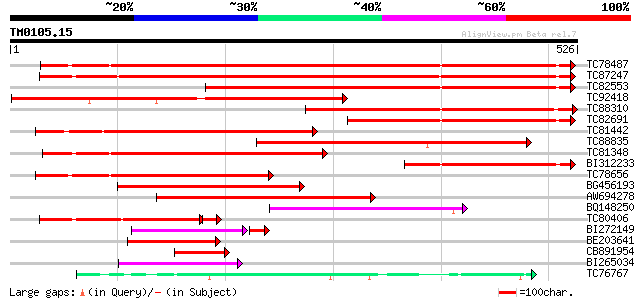
(526 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78487 similar to PIR|A84663|A84663 probable beta-ketoacyl-CoA ... 646 0.0
TC87247 similar to GP|9714501|emb|CAC01441.1 putative fatty acid... 596 e-171
TC82553 similar to PIR|T00951|T00951 probable 3-oxoacyl-[acyl-ca... 532 e-151
TC92418 similar to GP|3551960|gb|AAC34858.1| senescence-associat... 440 e-124
TC88310 similar to PIR|F86327|F86327 protein F18O14.21 [imported... 365 e-101
TC82691 similar to PIR|A84663|A84663 probable beta-ketoacyl-CoA ... 314 5e-86
TC81442 similar to PIR|F86327|F86327 protein F18O14.21 [imported... 265 2e-71
TC88835 similar to GP|16226847|gb|AAL16279.1 At1g07720/F24B9_16 ... 244 4e-65
TC81348 similar to GP|17979464|gb|AAL50069.1 At1g68530/T26J14_10... 242 2e-64
BI312233 homologue to GP|17979464|gb At1g68530/T26J14_10 {Arabid... 228 4e-60
TC78656 similar to GP|18447765|gb|AAL67993.1 fiddlehead-like pro... 223 2e-58
BG456193 similar to PIR|A84663|A846 probable beta-ketoacyl-CoA s... 178 5e-45
AW694278 similar to PIR|T48449|T48 fatty acid elongase-like prot... 174 1e-43
BQ148250 similar to GP|21536949|gb putative fatty acid elongase ... 154 6e-38
TC80406 weakly similar to GP|9714501|emb|CAC01441.1 putative fat... 103 3e-25
BI272149 weakly similar to GP|9714501|emb| putative fatty acid e... 86 4e-22
BE203641 weakly similar to PIR|F86141|F861 protein T25K16.11 [im... 80 3e-15
CB891954 similar to PIR|A84663|A846 probable beta-ketoacyl-CoA s... 69 6e-12
BI265034 similar to GP|21536949|gb putative fatty acid elongase ... 55 5e-08
TC76767 homologue to SP|P30074|CHS2_MEDSA Chalcone synthase 2 (E... 55 7e-08
>TC78487 similar to PIR|A84663|A84663 probable beta-ketoacyl-CoA synthase
[imported] - Arabidopsis thaliana, partial (97%)
Length = 1999
Score = 646 bits (1666), Expect = 0.0
Identities = 323/497 (64%), Positives = 392/497 (77%)
Frame = +1
Query: 29 RRLPDFLQTVKLKYVKLGYGYTCNAATVLIFTFIFPLILFLLLQLTDLKLIHLSELWSDH 88
R LPDF ++VKLKYVKLGY Y ++ F+ PL++ + QL+ L + ++W +
Sbjct: 280 RILPDFKKSVKLKYVKLGYHYLITHG---MYLFLSPLVVLIAAQLSTFSLNDIYDIWENL 450
Query: 89 TVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYLVDFACYKPEKERKISVDSFLKMTEETGG 148
+L + S +L+FL Y+ R PVYLV+F+CYKPE+ RK + F+ + +G
Sbjct: 451 QYNL-VSVIICSTLLVFLSTFYFMTRPRPVYLVNFSCYKPEESRKCTKRIFMDHSRASGF 627
Query: 149 FEEETLQFQRRISMKAGLGDETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALFAKTG 208
F EE L FQR+I ++GLG+ TYLP + S PPN MKEAR EAEAVMFGA+D LF+KT
Sbjct: 628 FTEENLDFQRKILERSGLGENTYLPEAVLSIPPNPSMKEARKEAEAVMFGAIDELFSKTT 807
Query: 209 VNPKDIDILVVNCSLFNPTPSLSAMIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKDLL 268
V PKDI IL+VNCSLF PTPSLSAMI+NHYKLR NI SYNLGGMGCSAG+ISIDLAK+LL
Sbjct: 808 VKPKDIGILIVNCSLFCPTPSLSAMIINHYKLRGNIMSYNLGGMGCSAGIISIDLAKELL 987
Query: 269 KANPNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSNKSSDRAKSKYELVH 328
+ +PNSYA+VVS ENITLNWY GNDRS L+ NC+FRMGGAA+LLSN+SSDR +SKY+LVH
Sbjct: 988 QVHPNSYALVVSMENITLNWYPGNDRSKLVSNCLFRMGGAAILLSNRSSDRRRSKYQLVH 1167
Query: 329 TVRTHKGSDDKHYNCVYQMEDEKGKVGVCLARELMAVAGDALKTNITTLGPLVLPFMEQF 388
TVRT+KG+DDK ++CV Q ED+ GKVGV L+++LMAVAGDALKTNITTLGPLVLP EQ
Sbjct: 1168TVRTNKGADDKCFSCVTQEEDDNGKVGVTLSKDLMAVAGDALKTNITTLGPLVLPTSEQL 1347
Query: 389 MFFVSLVRRKMTFLKRSKPYIPDFKLAFEHFCIHAGGRAVLDEMQKNLELSEWHMEPSRM 448
+FF +LV +K+ F + KPYIPDFKLAFEHFCIHAGGRAVLDE++KNL+LS WHMEPSRM
Sbjct: 1348LFFATLVGKKL-FKMKIKPYIPDFKLAFEHFCIHAGGRAVLDELEKNLKLSPWHMEPSRM 1524
Query: 449 TLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVWQIAFGSGFKCNSAVWKAVRDMPELGDW 508
TL+RFGNTSSSSLWYELAYTEA GR+ KGDR WQIAFGSGFKCNSAVWKA+R + +
Sbjct: 1525TLYRFGNTSSSSLWYELAYTEANGRIKKGDRTWQIAFGSGFKCNSAVWKALRTINPAKE- 1701
Query: 509 RGNPWDDSIDKYPVNVP 525
NPW D I ++PV+VP
Sbjct: 1702-KNPWIDEIHQFPVDVP 1749
>TC87247 similar to GP|9714501|emb|CAC01441.1 putative fatty acid elongase
{Zea mays}, partial (91%)
Length = 2102
Score = 596 bits (1536), Expect = e-171
Identities = 304/499 (60%), Positives = 376/499 (74%), Gaps = 1/499 (0%)
Frame = +3
Query: 28 RRRLPDFLQTVKLKYVKLGYGYTCNAATVLIFTFIFPLILFLLLQLTDLKLIHLSELWSD 87
RR+LP+FL +V+LKYVKLGY Y + A L+ + PL L+ + + +L+ +
Sbjct: 222 RRKLPNFLLSVRLKYVKLGYHYLISKAMYLL---LIPLFGAASAHLSTISYHVIIQLYEN 392
Query: 88 HTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYLVDFACYKPEKERKISVDSFLKMTEETG 147
+L + T+ S +++FL LY+ R VYLVDFACYKP++E + + F+ +E TG
Sbjct: 393 LKFNLVSVTLCTS-LMVFLVTLYFMSRPRGVYLVDFACYKPKQEYTCTREIFMNRSELTG 569
Query: 148 GFEEETLQFQRRISMKAGL-GDETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALFAK 206
F +E L FQ++I K + + P I +L + FGA+D L K
Sbjct: 570 TFSDENLAFQKKILEKVWFRSKDLFAPSNIECSTKSLYG*SKERSWRKLWFGAIDELLEK 749
Query: 207 TGVNPKDIDILVVNCSLFNPTPSLSAMIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKD 266
TGV KDI ILVVNCSLFNPTPSLSAMIVNHYKLR N++SYNLGGMGCSAGLISIDLAK
Sbjct: 750 TGVKAKDIGILVVNCSLFNPTPSLSAMIVNHYKLRGNVQSYNLGGMGCSAGLISIDLAKQ 929
Query: 267 LLKANPNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSNKSSDRAKSKYEL 326
LL+ +PNSYA+VVS ENITLNWYFGNDRSML+ NC+FRMGG+A+LLSNK+SD ++KY+L
Sbjct: 930 LLQVHPNSYALVVSMENITLNWYFGNDRSMLVSNCLFRMGGSAILLSNKASDSKRAKYQL 1109
Query: 327 VHTVRTHKGSDDKHYNCVYQMEDEKGKVGVCLARELMAVAGDALKTNITTLGPLVLPFME 386
+HTVRTHKG+DD+ Y CV+Q EDEK +GV L+++LMAVAG+ALKTNITTLGPLVLP E
Sbjct: 1110IHTVRTHKGADDRSYGCVFQEEDEKKSIGVSLSKDLMAVAGEALKTNITTLGPLVLPMSE 1289
Query: 387 QFMFFVSLVRRKMTFLKRSKPYIPDFKLAFEHFCIHAGGRAVLDEMQKNLELSEWHMEPS 446
Q +FF +LV RK F + KPYIPDFKLAFEHFCIHAGGRAVLDE++KNL+LS+WHMEPS
Sbjct: 1290QLLFFATLVARK-AFKMKIKPYIPDFKLAFEHFCIHAGGRAVLDELEKNLDLSDWHMEPS 1466
Query: 447 RMTLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVWQIAFGSGFKCNSAVWKAVRDMPELG 506
RMTL+RFGNTSSSSLWYELAYTEAKGR+ KGDR WQIAFGSGFKCNSAVW+A+R +
Sbjct: 1467RMTLNRFGNTSSSSLWYELAYTEAKGRIRKGDRTWQIAFGSGFKCNSAVWRALRTVDPAK 1646
Query: 507 DWRGNPWDDSIDKYPVNVP 525
+ NPW D I ++PV+VP
Sbjct: 1647E--KNPWMDEIHEFPVHVP 1697
>TC82553 similar to PIR|T00951|T00951 probable
3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41)
F20D22.1 - Arabidopsis thaliana, partial (65%)
Length = 1343
Score = 532 bits (1371), Expect = e-151
Identities = 262/344 (76%), Positives = 297/344 (86%)
Frame = +3
Query: 182 NLCMKEARLEAEAVMFGAMDALFAKTGVNPKDIDILVVNCSLFNPTPSLSAMIVNHYKLR 241
N CM EAR EAE VMFGA+D + KTGV KDI ILVVNCSLFNPTPSLSAMIVNHYKLR
Sbjct: 18 NPCMAEARKEAEEVMFGAIDEVLQKTGVKAKDIGILVVNCSLFNPTPSLSAMIVNHYKLR 197
Query: 242 TNIKSYNLGGMGCSAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGNDRSMLLCNC 301
NI SYNLGGMGCSAGLISIDLAK LL+ +PNSYA+VVS ENITLNWYFGNDRSML+ NC
Sbjct: 198 GNILSYNLGGMGCSAGLISIDLAKQLLQVHPNSYALVVSMENITLNWYFGNDRSMLVPNC 377
Query: 302 IFRMGGAAVLLSNKSSDRAKSKYELVHTVRTHKGSDDKHYNCVYQMEDEKGKVGVCLARE 361
+FRMGGAAVLLSNK DR +SKY+LVHTVRTHKG+D+K Y CV+Q ED+ +VGV L+++
Sbjct: 378 LFRMGGAAVLLSNKPRDRLRSKYQLVHTVRTHKGADNKSYGCVFQEEDDTKQVGVSLSKD 557
Query: 362 LMAVAGDALKTNITTLGPLVLPFMEQFMFFVSLVRRKMTFLKRSKPYIPDFKLAFEHFCI 421
LMAVAG+ALKTNITTLGPLVLP EQ +FF +LV RK+ F + KPYIPDFKLAFEHFCI
Sbjct: 558 LMAVAGEALKTNITTLGPLVLPMSEQLLFFGTLVARKI-FKMKIKPYIPDFKLAFEHFCI 734
Query: 422 HAGGRAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVW 481
HAGGRAVLDE++KNL+LS+WHMEPSRMTL+RFGNTSSSSLWYELAYTEAKGR+ KGDR W
Sbjct: 735 HAGGRAVLDELEKNLDLSDWHMEPSRMTLNRFGNTSSSSLWYELAYTEAKGRIKKGDRTW 914
Query: 482 QIAFGSGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKYPVNVP 525
QIAFGSGFKCNSAVWKA++ + + NPW D I ++PV+VP
Sbjct: 915 QIAFGSGFKCNSAVWKALKTINPAKE--KNPWIDEIHEFPVHVP 1040
>TC92418 similar to GP|3551960|gb|AAC34858.1| senescence-associated protein
15 {Hemerocallis hybrid cultivar}, partial (35%)
Length = 1034
Score = 440 bits (1131), Expect = e-124
Identities = 234/321 (72%), Positives = 256/321 (78%), Gaps = 9/321 (2%)
Frame = +2
Query: 2 DSIDMDKERLTAEMDFKDSTSAVIKIRRRLPDFLQTVKLKYVKLGYGYTCNAATVLIFTF 61
+SIDMDKERLTAEMDFKDSTSAVIKIRRRLPDFLQTVKLKYVKLGYGY+CNA T+LIFTF
Sbjct: 59 NSIDMDKERLTAEMDFKDSTSAVIKIRRRLPDFLQTVKLKYVKLGYGYSCNATTILIFTF 238
Query: 62 IFPLILFLLLQ--LTDLKLIHLSELWSDHTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVY 119
+ PL+LFL +Q LTDLKL LS+L +HTV LDTDT GS LLFLFGLY+AKRS P+Y
Sbjct: 239 VVPLLLFLTVQFQLTDLKLHRLSQLLLEHTVQLDTDTAIGSIFLLFLFGLYYAKRSPPIY 418
Query: 120 LVDFACYKPEKERKI-------SVDSFLKMTEETGGFEEETLQFQRRISMKAGLGDETYL 172
LVDFACYKPE + +V ++ R + MK E
Sbjct: 419 LVDFACYKPEN*AENCS*IFR*NVRR*WRIRRRNTSISTPHFVKSRSLVMKLPCQTE*CQ 598
Query: 173 PRGITSRPPNLCMKEARLEAEAVMFGAMDALFAKTGVNPKDIDILVVNCSLFNPTPSLSA 232
NLCMKEARLEAE+VMFG++DALFAKTGVNP+DIDILVVNCSLFNPTPSLSA
Sbjct: 599 VH------QNLCMKEARLEAESVMFGSLDALFAKTGVNPRDIDILVVNCSLFNPTPSLSA 760
Query: 233 MIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGN 292
MIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKDLLKANPNSYAVV+STEN+TLNWYFGN
Sbjct: 761 MIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKDLLKANPNSYAVVLSTENLTLNWYFGN 940
Query: 293 DRSMLLCNCIFRMGGAAVLLS 313
DRSMLL NCIFRMGGAA+LLS
Sbjct: 941 DRSMLLSNCIFRMGGAAILLS 1003
>TC88310 similar to PIR|F86327|F86327 protein F18O14.21 [imported] -
Arabidopsis thaliana, partial (47%)
Length = 995
Score = 365 bits (936), Expect = e-101
Identities = 174/252 (69%), Positives = 212/252 (84%)
Frame = +3
Query: 275 YAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSNKSSDRAKSKYELVHTVRTHK 334
YAVVVSTENIT NWYFGN +SML+ NC+FR+GGAAVLLSNK DR+++KY+LVH VRTHK
Sbjct: 3 YAVVVSTENITQNWYFGNKKSMLIPNCLFRVGGAAVLLSNKGCDRSRAKYKLVHVVRTHK 182
Query: 335 GSDDKHYNCVYQMEDEKGKVGVCLARELMAVAGDALKTNITTLGPLVLPFMEQFMFFVSL 394
G+DDK + CVYQ +D+ GK GV L+++LMA+AG ALKTNITTLGPLVLP EQ +FF +L
Sbjct: 183 GADDKAFKCVYQEQDDVGKTGVSLSKDLMAIAGGALKTNITTLGPLVLPVSEQLLFFTTL 362
Query: 395 VRRKMTFLKRSKPYIPDFKLAFEHFCIHAGGRAVLDEMQKNLELSEWHMEPSRMTLHRFG 454
V +K F ++KPYIPDFKLAFEHFCIHAGGRAV+DE++KNL+L H+E SRMTLHRFG
Sbjct: 363 VIKKW-FNAKTKPYIPDFKLAFEHFCIHAGGRAVIDELEKNLQLMPDHVEASRMTLHRFG 539
Query: 455 NTSSSSLWYELAYTEAKGRVGKGDRVWQIAFGSGFKCNSAVWKAVRDMPELGDWRGNPWD 514
NTSSSS+WYELAY EAKGR+ KG+R+WQIAFGSGFKCNSAVW+A++ + +PW+
Sbjct: 540 NTSSSSIWYELAYIEAKGRMRKGNRIWQIAFGSGFKCNSAVWQAMKHVKAS---PMSPWE 710
Query: 515 DSIDKYPVNVPS 526
D ID+YPV + S
Sbjct: 711 DCIDRYPVEIVS 746
>TC82691 similar to PIR|A84663|A84663 probable beta-ketoacyl-CoA synthase
[imported] - Arabidopsis thaliana, partial (41%)
Length = 823
Score = 314 bits (805), Expect = 5e-86
Identities = 152/212 (71%), Positives = 181/212 (84%)
Frame = +2
Query: 314 NKSSDRAKSKYELVHTVRTHKGSDDKHYNCVYQMEDEKGKVGVCLARELMAVAGDALKTN 373
+KSSDR +SKY+L+ TVRT+K SDDK ++CV Q ED GK+GV L+++LMAVAGDALKTN
Sbjct: 14 HKSSDRRRSKYQLITTVRTNKASDDKCFSCVTQEEDANGKIGVTLSKDLMAVAGDALKTN 193
Query: 374 ITTLGPLVLPFMEQFMFFVSLVRRKMTFLKRSKPYIPDFKLAFEHFCIHAGGRAVLDEMQ 433
ITTLGPLVLP EQ +FF +LV +K+ F + KPYIPDFKLAFEHFCIHAGGRAVLDE++
Sbjct: 194 ITTLGPLVLPTSEQLLFFTTLVGKKL-FKMKIKPYIPDFKLAFEHFCIHAGGRAVLDELE 370
Query: 434 KNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVWQIAFGSGFKCNS 493
KNL+LS WHMEPSRMTL+RFGNTSSSSLWYELAYTEAKGR+ KGDR WQIAFGSGFKCNS
Sbjct: 371 KNLQLSPWHMEPSRMTLYRFGNTSSSSLWYELAYTEAKGRIKKGDRTWQIAFGSGFKCNS 550
Query: 494 AVWKAVRDMPELGDWRGNPWDDSIDKYPVNVP 525
AVWKA+R + + + +PW D I ++PV+VP
Sbjct: 551 AVWKALRTINPVKE--KSPWIDEIGQFPVDVP 640
>TC81442 similar to PIR|F86327|F86327 protein F18O14.21 [imported] -
Arabidopsis thaliana, partial (47%)
Length = 915
Score = 265 bits (678), Expect = 2e-71
Identities = 137/261 (52%), Positives = 182/261 (69%)
Frame = +2
Query: 25 IKIRRRLPDFLQTVKLKYVKLGYGYTCNAATVLIFTFIFPLILFLLLQLTDLKLIHLSEL 84
I RRLPDFLQ+V LKYVKLGY Y T L+ + PL+ +++Q++ + + +L
Sbjct: 140 IHTNRRLPDFLQSVNLKYVKLGYHYLI---THLLTLCLIPLMSVIIIQVSQMNPNEIHQL 310
Query: 85 WSDHTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYLVDFACYKPEKERKISVDSFLKMTE 144
W H + +T SA+L+F +Y R +YL+DF+CYKP + F++ ++
Sbjct: 311 WL-HLKYNLVSIITCSAILVFGSTVYIMTRPRSIYLIDFSCYKPPSNLAVKFTKFIQHSK 487
Query: 145 ETGGFEEETLQFQRRISMKAGLGDETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALF 204
G F+E +L+FQR+I ++GLG++TYLP + PP CM AR EAE VM+GA+D LF
Sbjct: 488 LKGDFDESSLEFQRKILERSGLGEDTYLPEAMHKIPPTPCMASAREEAEQVMYGALDNLF 667
Query: 205 AKTGVNPKDIDILVVNCSLFNPTPSLSAMIVNHYKLRTNIKSYNLGGMGCSAGLISIDLA 264
A T + PKDI ILVVNCSLFNPTPSLSAMIVN YKLR NI+S+NLGGMGCSAG+I+IDLA
Sbjct: 668 ANTKIKPKDIGILVVNCSLFNPTPSLSAMIVNKYKLRGNIRSFNLGGMGCSAGVIAIDLA 847
Query: 265 KDLLKANPNSYAVVVSTENIT 285
KD+L+ + N+YAVV STENIT
Sbjct: 848 KDMLQVHRNTYAVVDSTENIT 910
>TC88835 similar to GP|16226847|gb|AAL16279.1 At1g07720/F24B9_16
{Arabidopsis thaliana}, partial (51%)
Length = 788
Score = 244 bits (624), Expect = 4e-65
Identities = 124/258 (48%), Positives = 173/258 (66%), Gaps = 3/258 (1%)
Frame = +3
Query: 230 LSAMIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKDLLKANPNSYAVVVSTENITLNWY 289
LS+ I+N YK+R ++K YNL MGCSA LIS+D+ K++ K+ N A++V++E++ NWY
Sbjct: 3 LSSRIINRYKMRHDVKVYNLTAMGCSASLISLDIVKNIFKSQRNKLALLVTSESLCPNWY 182
Query: 290 FGNDRSMLLCNCIFRMGGAAVLLSNKSSDRAKSKYELVHTVRTHKGSDDKHYNCVYQMED 349
GND+SM+L NC+FR GG AVLL+NK S + +S +L VRTH G+ D Y C Q ED
Sbjct: 183 TGNDKSMILANCLFRSGGCAVLLTNKRSLKNRSILKLKCLVRTHHGARDDSYTCCTQKED 362
Query: 350 EKGKVGVCLARELMAVAGDALKTNITTLGPLVLPFME--QFMFFVSLVRRKMTFLKRSKP 407
E+G++G+ L + L A A N+ + P +LP E +F+F L + K T
Sbjct: 363 EQGRLGIHLGKTLPKAATRAFVDNLRVISPKILPTKEILRFLFVTLLKKLKKTSGGTKST 542
Query: 408 YIP-DFKLAFEHFCIHAGGRAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELA 466
P +FK +HFC+H GG+AV+D + +L+LSE+ +EP+RMTLHRFGNTS+SSLWY L
Sbjct: 543 KSPLNFKTGVDHFCLHTGGKAVIDGIGMSLDLSEYDLEPARMTLHRFGNTSASSLWYVLX 722
Query: 467 YTEAKGRVGKGDRVWQIA 484
Y EAK R+ KGDRV I+
Sbjct: 723 YMEAKKRLKKGDRVXMIS 776
>TC81348 similar to GP|17979464|gb|AAL50069.1 At1g68530/T26J14_10
{Arabidopsis thaliana}, partial (53%)
Length = 906
Score = 242 bits (618), Expect = 2e-64
Identities = 129/265 (48%), Positives = 181/265 (67%)
Frame = +2
Query: 31 LPDFLQTVKLKYVKLGYGYTCNAATVLIFTFIFPLILFLLLQLTDLKLIHLSELWSDHTV 90
LPDF +VKLKYVKLGY Y N L + P+++ + ++L L + LW+ +
Sbjct: 92 LPDFSSSVKLKYVKLGYQYLVNHIITLT---LIPIMIGVSIELIRLGPNEILNLWNSLNL 262
Query: 91 HLDTDTVTGSAVLLFLFGLYWAKRSAPVYLVDFACYKPEKERKISVDSFLKMTEETGGFE 150
+L + S +++F+ +Y+ + +YLVD+AC+KP ++ +F++ +
Sbjct: 263 NL-VQILCSSFLVIFIATVYFMSKPRTIYLVDYACFKPPVTCRVPFATFMEHSRLILKNN 439
Query: 151 EETLQFQRRISMKAGLGDETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALFAKTGVN 210
++++FQ RI ++GLG+ET LP I PP M+ AR EAE V+F AMD+LF KTG+
Sbjct: 440 PKSVEFQMRILERSGLGEETCLPPSIHYIPPKPTMESARGEAELVIFSAMDSLFEKTGLK 619
Query: 211 PKDIDILVVNCSLFNPTPSLSAMIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKDLLKA 270
PKDIDIL+VNCSLF+PTPSLSAM++N YKLR+NI+S+NL GMGCSAGLISIDLA+DLL+
Sbjct: 620 PKDIDILIVNCSLFSPTPSLSAMVINKYKLRSNIRSFNLSGMGCSAGLISIDLARDLLQV 799
Query: 271 NPNSYAVVVSTENITLNWYFGNDRS 295
+PNS AVVVSTE IT N+Y G +S
Sbjct: 800 HPNSNAVVVSTEIITPNYYQGK*KS 874
>BI312233 homologue to GP|17979464|gb At1g68530/T26J14_10 {Arabidopsis
thaliana}, partial (32%)
Length = 744
Score = 228 bits (581), Expect = 4e-60
Identities = 111/159 (69%), Positives = 129/159 (80%)
Frame = +1
Query: 367 GDALKTNITTLGPLVLPFMEQFMFFVSLVRRKMTFLKRSKPYIPDFKLAFEHFCIHAGGR 426
G ALK+NITT+GPLVLP EQ +F ++L+ RK+ F + KPYIPDFK AFEHFCIHAGGR
Sbjct: 1 GKALKSNITTMGPLVLPASEQLLFLITLIGRKI-FNPKWKPYIPDFKQAFEHFCIHAGGR 177
Query: 427 AVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVWQIAFG 486
AV+DE+QKNL+LS H+E SRMTLHRFGNTSSSSLWYEL Y E+KGR+ KGDRVWQIAFG
Sbjct: 178 AVIDELQKNLQLSTEHVEASRMTLHRFGNTSSSSLWYELNYIESKGRMKKGDRVWQIAFG 357
Query: 487 SGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKYPVNVP 525
SGFKCNSAVWK R + D PW D ID+YPV++P
Sbjct: 358 SGFKCNSAVWKCNRSIKTPID---GPWTDCIDRYPVHIP 465
>TC78656 similar to GP|18447765|gb|AAL67993.1 fiddlehead-like protein
{Gossypium hirsutum}, partial (46%)
Length = 806
Score = 223 bits (567), Expect = 2e-58
Identities = 111/223 (49%), Positives = 154/223 (68%), Gaps = 3/223 (1%)
Frame = +2
Query: 25 IKIRRRLPDFLQTVKLKYVKLGYGYTCNAATVLIFTFIFPLILFLL-LQLTDLKLIHL-S 82
+++RRRLPDFLQ+V LKYVKLGY Y N ++ F P++L + ++ L L
Sbjct: 149 VRVRRRLPDFLQSVNLKYVKLGYHYLINHG---VYLFTIPILLVVFSAEVGSLSKEDLWK 319
Query: 83 ELWSDHTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYLVDFACYKPEKERKISVDSFLKM 142
++W D T L ++ AV +F F LY+ R P+YL+DFACY+P+ E K+S + +++
Sbjct: 320 KIWEDATYDL-ASVLSSLAVFVFTFTLYFMSRPRPIYLIDFACYQPDDELKVSREQLIEL 496
Query: 143 TEETGGFEEETLQFQRRISMKAGLGDETYLPRGITSRPPNLC-MKEARLEAEAVMFGAMD 201
++G F+E +L+FQ+RI M +G+GDETY+PR + S N MKE R EA VMFGA+D
Sbjct: 497 ARKSGKFDEGSLEFQKRIVMSSGIGDETYIPRSVISSSENTATMKEGRAEASMVMFGALD 676
Query: 202 ALFAKTGVNPKDIDILVVNCSLFNPTPSLSAMIVNHYKLRTNI 244
LF KTG+ PKD+ +LVVNCS+FNPTPSLSAMI+NHYK+R NI
Sbjct: 677 ELFEKTGIRPKDVGVLVVNCSIFNPTPSLSAMIINHYKMRGNI 805
>BG456193 similar to PIR|A84663|A846 probable beta-ketoacyl-CoA synthase
[imported] - Arabidopsis thaliana, partial (16%)
Length = 643
Score = 178 bits (451), Expect = 5e-45
Identities = 86/173 (49%), Positives = 120/173 (68%)
Frame = +1
Query: 101 AVLLFLFGLYWAKRSAPVYLVDFACYKPEKERKISVDSFLKMTEETGGFEEETLQFQRRI 160
A + ++ Y K S VYLVDFACYKP S + F+K T+ G F++E++ FQ++I
Sbjct: 124 ASISYIVTFYQKKCSKKVYLVDFACYKPFPNGICSKELFIKQTKSGGNFKDESIDFQKKI 303
Query: 161 SMKAGLGDETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALFAKTGVNPKDIDILVVN 220
++G GD+TY+P + P N+ + EAR E E+V+FGA++ L KT + +DI+IL+ N
Sbjct: 304 LDRSGFGDKTYVPESLLKIPQNISIVEARKETESVIFGAINDLLLKTKMKAEDIEILITN 483
Query: 221 CSLFNPTPSLSAMIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKDLLKANPN 273
CS+FNP PSLSAM+VNH+KL+ I YNL GMGCSA LI+IDLAK LL+ + N
Sbjct: 484 CSIFNPVPSLSAMVVNHFKLKHTILCYNLSGMGCSAXLIAIDLAKQLLQVHQN 642
>AW694278 similar to PIR|T48449|T48 fatty acid elongase-like protein -
Arabidopsis thaliana, partial (39%)
Length = 641
Score = 174 bits (440), Expect = 1e-43
Identities = 86/204 (42%), Positives = 128/204 (62%), Gaps = 1/204 (0%)
Frame = +3
Query: 137 DSFLKMTEETGGFEEETLQFQRRISMKAGLGDETYLPRGITS-RPPNLCMKEARLEAEAV 195
DS K+ E +F + + +G+G+ TY PR + R +K+ E + +
Sbjct: 6 DSAAKIVMRNKNLGLEEFRFLLKTMVSSGIGENTYCPRNVLEGRETCPTLKDTYEEIDEI 185
Query: 196 MFGAMDALFAKTGVNPKDIDILVVNCSLFNPTPSLSAMIVNHYKLRTNIKSYNLGGMGCS 255
MF +D LF KT +P +IDILVVN SLF+P PSL+A I+N YK+R ++K +NL GMGCS
Sbjct: 186 MFDTLDNLFKKTCFSPSEIDILVVNVSLFSPAPSLTARIINRYKMREDVKVFNLAGMGCS 365
Query: 256 AGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSNK 315
A +++IDL + L K NS +VVSTE++ +WY G D+ M+L NC+FR GG ++L +NK
Sbjct: 366 ASVVAIDLVQQLFKTYENSLGIVVSTEDLGSHWYCGKDKKMMLSNCLFRAGGCSMLFTNK 545
Query: 316 SSDRAKSKYELVHTVRTHKGSDDK 339
+ + ++ +L H RT GSDD+
Sbjct: 546 TELKDRAILKLKHMERTQYGSDDE 617
>BQ148250 similar to GP|21536949|gb putative fatty acid elongase {Arabidopsis
thaliana}, partial (36%)
Length = 647
Score = 154 bits (390), Expect = 6e-38
Identities = 79/194 (40%), Positives = 116/194 (59%), Gaps = 11/194 (5%)
Frame = +1
Query: 242 TNIKSYNLGGMGCSAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGNDRSMLLCNC 301
T++K YNL GMGCSA LIS+D+ K++ K+ N A++V++E+++ NWY GNDRSM+L NC
Sbjct: 61 TDVKVYNLTGMGCSASLISLDIVKNIFKSQRNKLALLVTSESLSPNWYTGNDRSMILANC 240
Query: 302 IFRMGGAAVLLSNKSSDRAKSKYELVHTVRTHKGSDDKHYNCVYQMEDEKGKVGVCLARE 361
+FR GG ++LL+NK S + +S +L VRTH G+ + YNC Q EDE+G++G L +
Sbjct: 241 LFRSGGCSILLTNKRSLKNRSILKLKCLVRTHHGAREDSYNCCIQKEDEQGRLGFHLGKT 420
Query: 362 LMAVAGDALKTNITTLGPLVLPFMEQFMFFVSLVRRKMTF-LKRSKPYIP---------- 410
L A N+ + P +LP E F + L+ K+ SK Y
Sbjct: 421 LPKAATKVFVDNLRVISPKILPTRELLRFLLVLLFNKLKIATSSSKSYSGGATKSTKSPL 600
Query: 411 DFKLAFEHFCIHAG 424
+FK +HFC+H G
Sbjct: 601 NFKTGVDHFCLHTG 642
>TC80406 weakly similar to GP|9714501|emb|CAC01441.1 putative fatty acid
elongase {Zea mays}, partial (30%)
Length = 726
Score = 103 bits (258), Expect(2) = 3e-25
Identities = 59/153 (38%), Positives = 93/153 (60%)
Frame = +3
Query: 28 RRRLPDFLQTVKLKYVKLGYGYTCNAATVLIFTFIFPLILFLLLQLTDLKLIHLSELWSD 87
R +LP+FL +VKLKYVKLGY Y + A L+ + PL+ + L+ + L +++ +
Sbjct: 228 RSKLPNFLISVKLKYVKLGYHYLISNAMYLV---LIPLLGVASVHLSTFSIKDLIQIYEN 398
Query: 88 HTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYLVDFACYKPEKERKISVDSFLKMTEETG 147
+ + + S V+ FL LY+ R +YLVDFACYKP+K+ +++ + F++ + T
Sbjct: 399 LKFNFVSMLICTSLVV-FLATLYFMSRPRGIYLVDFACYKPQKDLQVTREIFVERSILTK 575
Query: 148 GFEEETLQFQRRISMKAGLGDETYLPRGITSRP 180
F EETL FQ++I ++GLG +TYLP I P
Sbjct: 576 AFTEETLSFQKKILERSGLGQKTYLPPXIMRVP 674
Score = 29.6 bits (65), Expect(2) = 3e-25
Identities = 13/17 (76%), Positives = 13/17 (76%)
Frame = +2
Query: 180 PPNLCMKEARLEAEAVM 196
PPN CM EAR EAE VM
Sbjct: 671 PPNPCMAEARKEAEXVM 721
>BI272149 weakly similar to GP|9714501|emb| putative fatty acid elongase {Zea
mays}, partial (15%)
Length = 631
Score = 85.9 bits (211), Expect(2) = 4e-22
Identities = 44/107 (41%), Positives = 61/107 (56%)
Frame = +2
Query: 114 RSAPVYLVDFACYKPEKERKISVDSFLKMTEETGGFEEETLQFQRRISMKAGLGDETYLP 173
R +YLVDFAC+KP+ + + L+ G EE+ +I ++G+G TY+P
Sbjct: 209 RPNKIYLVDFACHKPKNNCLCTKEMLLERANNFGFLNEESFILIHKILERSGVGPTTYVP 388
Query: 174 RGITSRPPNLCMKEARLEAEAVMFGAMDALFAKTGVNPKDIDILVVN 220
+ PP L + EAR E+ V+FGA+D L KT V KDI ILVVN
Sbjct: 389 EALLEMPPRLTLDEARNESNLVLFGAVDELLEKTKVEAKDIGILVVN 529
Score = 37.0 bits (84), Expect(2) = 4e-22
Identities = 16/19 (84%), Positives = 17/19 (89%)
Frame = +3
Query: 223 LFNPTPSLSAMIVNHYKLR 241
LFNPTPSLS IVNHYKL+
Sbjct: 537 LFNPTPSLSDTIVNHYKLK 593
>BE203641 weakly similar to PIR|F86141|F861 protein T25K16.11 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 403
Score = 79.7 bits (195), Expect = 3e-15
Identities = 37/86 (43%), Positives = 54/86 (62%)
Frame = +1
Query: 110 YWAKRSAPVYLVDFACYKPEKERKISVDSFLKMTEETGGFEEETLQFQRRISMKAGLGDE 169
Y K S +YLVDF+CYKP S + F++ + G F +E++ FQ++I K+G GD+
Sbjct: 145 YLKKCSKNIYLVDFSCYKPFPRSICSKELFIETSRCGGHFRDESIDFQKKIMEKSGFGDK 324
Query: 170 TYLPRGITSRPPNLCMKEARLEAEAV 195
TY+P + + PPN+C AR E EAV
Sbjct: 325 TYVPENLLNIPPNICTNVARKETEAV 402
>CB891954 similar to PIR|A84663|A846 probable beta-ketoacyl-CoA synthase
[imported] - Arabidopsis thaliana, partial (15%)
Length = 402
Score = 68.6 bits (166), Expect = 6e-12
Identities = 33/51 (64%), Positives = 39/51 (75%)
Frame = +1
Query: 154 LQFQRRISMKAGLGDETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALF 204
L FQR+I ++GLG+ TYLP + S PPN MKEAR EAEAVMFGA+D LF
Sbjct: 205 LDFQRKILERSGLGENTYLPEAVLSIPPNPSMKEARKEAEAVMFGAIDELF 357
Score = 29.6 bits (65), Expect = 3.0
Identities = 13/19 (68%), Positives = 16/19 (83%)
Frame = +1
Query: 29 RRLPDFLQTVKLKYVKLGY 47
R LPDF ++VKLKYVKL +
Sbjct: 157 RILPDFKKSVKLKYVKLDF 213
>BI265034 similar to GP|21536949|gb putative fatty acid elongase {Arabidopsis
thaliana}, partial (23%)
Length = 427
Score = 55.5 bits (132), Expect = 5e-08
Identities = 27/116 (23%), Positives = 57/116 (48%), Gaps = 1/116 (0%)
Frame = +3
Query: 102 VLLFLFGLYWAKRSAPVYLVDFACYKPEKERKISVDSFLKMTEETGGFEEETLQFQRRIS 161
+ ++ LY KR Y++D+ CYKP ++R + + K+ T + +F +
Sbjct: 78 ICFLIWKLYDQKRDQECYILDYQCYKPTQDRMLGTEFCGKIIRRTKNLGLDEYKFLLKAV 257
Query: 162 MKAGLGDETYLPRGI-TSRPPNLCMKEARLEAEAVMFGAMDALFAKTGVNPKDIDI 216
+ +G+G++TY PR + R + + + E E ++ L ++ ++P + DI
Sbjct: 258 VSSGIGEQTYAPRNVFEGRESSPTLNDGIXEMEEFFNDSIAKLLXRSAISPXEXDI 425
>TC76767 homologue to SP|P30074|CHS2_MEDSA Chalcone synthase 2 (EC 2.3.1.74)
(Naringenin-chalcone synthase 2). [Alfalfa] {Medicago
sativa}, complete
Length = 1436
Score = 55.1 bits (131), Expect = 7e-08
Identities = 102/449 (22%), Positives = 167/449 (36%), Gaps = 23/449 (5%)
Frame = +2
Query: 63 FPLILFLLLQLTDLKLIHLSELWSDHTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYLVD 122
F IL L+ D+K++ +SE+ + G A +L + + P V+
Sbjct: 83 FVQILVLIFATKDIKMVSVSEI-------RNAQRAEGPATILAI------GTANPANCVE 223
Query: 123 FACYKPEKERKISVDSFLKMTEETGGFEEETLQFQRRISMKAGLGDETYLPRGITSRPPN 182
+ Y D + K+T E + +FQR YL I P+
Sbjct: 224 QSTYP---------DFYFKITNSEHKTELKE-KFQRMCDKSMIKRRYMYLTEEILKENPS 373
Query: 183 LC--------MKEARLEAEAVMFGAMDALFA-KTGVNPKD-IDILVVNCSLFNPTPSLSA 232
+C ++ + E G A+ A K PK I L+V + P
Sbjct: 374 VCEYMAPSLDARQDMVVVEVPRLGKEAAVKAIKEWGQPKSKITHLIVCTTSGVDMPGADY 553
Query: 233 MIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGN 292
+ LR +K Y + GC AG + LAKDL + N + +VV +E + + +
Sbjct: 554 QLTKLLGLRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSEVTAVTFRGPS 733
Query: 293 DRSM--LLCNCIFRMGGAAVLL-SNKSSDRAKSKYELVHTVRT----HKGSDDKHYNCVY 345
D + L+ +F G AA+++ S+ + K +E+V T +T +G+ D H
Sbjct: 734 DTHLDSLVGQALFGDGAAALIVGSDPVPEIEKPIFEMVWTAQTIAPDSEGAIDGHLR--- 904
Query: 346 QMEDEKGKVGVCLARELMAVAGDALKTNITTLGPLVLPFMEQFMFFVSLVRRKMTFLKRS 405
E G L V+ + K + PL
Sbjct: 905 ----EAGLTFHLLKDVPGIVSKNITKALVEAFEPL------------------------- 997
Query: 406 KPYIPDFKLAFEHFCIHAGGRAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYEL 465
I D+ F + H GG A+LD++++ L L M +R L +GN SS+ + + L
Sbjct: 998 --GISDYNSIF--WIAHPGGPAILDQVEQKLALKPEKMNATREVLSEYGNMSSACVLFIL 1165
Query: 466 AYTEAKG------RVGKGDRVWQIAFGSG 488
K G+G W + FG G
Sbjct: 1166DEMRKKSTKDGLKTTGEG-LEWGVLFGFG 1249
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,526,590
Number of Sequences: 36976
Number of extensions: 261601
Number of successful extensions: 1540
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 1489
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1514
length of query: 526
length of database: 9,014,727
effective HSP length: 101
effective length of query: 425
effective length of database: 5,280,151
effective search space: 2244064175
effective search space used: 2244064175
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0105.15


