

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0104.7
(76 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
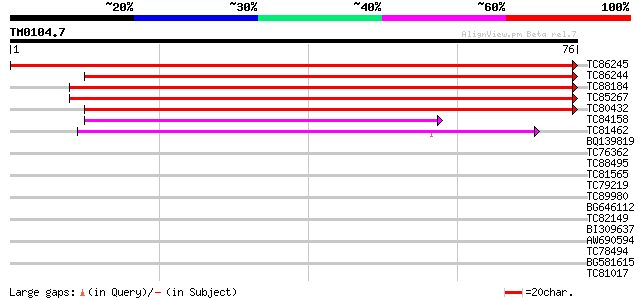
Score E
Sequences producing significant alignments: (bits) Value
TC86245 weakly similar to GP|9279671|dbj|BAB01228.1 transport in... 133 1e-32
TC86244 weakly similar to PIR|T48087|T48087 transport inhibitor ... 113 2e-26
TC88184 similar to GP|11935209|gb|AAG42020.1 putative transport ... 95 6e-21
TC85267 similar to GP|9279671|dbj|BAB01228.1 transport inhibitor... 91 1e-19
TC80432 similar to PIR|T52139|T52139 LRR-containing F-box protei... 58 6e-10
TC84158 similar to PIR|T52139|T52139 LRR-containing F-box protei... 43 3e-05
TC81462 similar to GP|21536497|gb|AAM60829.1 F-box protein famil... 39 3e-04
BQ139819 37 0.002
TC76362 similar to GP|10716949|gb|AAG21977.1 SKP1 interacting pa... 35 0.004
TC88495 similar to GP|15420162|gb|AAK97303.1 F-box containing pr... 35 0.007
TC81565 similar to GP|6041850|gb|AAF02159.1| unknown protein {Ar... 34 0.012
TC79219 similar to GP|15810000|gb|AAL06927.1 AT5g23340/MKD15_20 ... 33 0.027
TC89980 similar to PIR|D96512|D96512 hypothetical protein F2G19.... 32 0.035
BG646112 weakly similar to GP|10177368|dbj contains similarity t... 30 0.17
TC82149 weakly similar to PIR|H96695|H96695 hypothetical protein... 30 0.23
BI309637 similar to PIR|D86241|D86 protein T16B5.8 [imported] - ... 30 0.23
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 29 0.38
TC78494 28 0.50
BG581615 similar to GP|10177368|dbj contains similarity to heat ... 28 0.50
TC81017 weakly similar to GP|21632795|gb|AAL47185.1 phosphoinosi... 25 7.2
>TC86245 weakly similar to GP|9279671|dbj|BAB01228.1 transport inhibitor
response-like protein {Arabidopsis thaliana}, partial
(80%)
Length = 2260
Score = 133 bits (335), Expect = 1e-32
Identities = 64/76 (84%), Positives = 70/76 (91%)
Frame = +3
Query: 1 NILNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEI 60
N ++ S FPDEVLERVLGM+KSRKDRSSVSLVCKEWYNAERWSRR+VFIGNCY+VSPEI
Sbjct: 492 NNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEI 671
Query: 61 LTRRSSNIRSVTLKGK 76
LTRR NIRSVT+KGK
Sbjct: 672 LTRRFPNIRSVTMKGK 719
>TC86244 weakly similar to PIR|T48087|T48087 transport inhibitor response
protein TIR1 [imported] - Arabidopsis thaliana, partial
(48%)
Length = 2369
Score = 113 bits (282), Expect = 2e-26
Identities = 54/66 (81%), Positives = 61/66 (91%)
Frame = +1
Query: 11 DEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNIRS 70
DEVL++VLG VKSRKDR+S SLVCK+WYNAER SRR+VFIGNCYSV+PEILTRR NIRS
Sbjct: 250 DEVLKQVLGTVKSRKDRNSASLVCKQWYNAERLSRRNVFIGNCYSVTPEILTRRFPNIRS 429
Query: 71 VTLKGK 76
+TLKGK
Sbjct: 430 ITLKGK 447
>TC88184 similar to GP|11935209|gb|AAG42020.1 putative transport inhibitor
response TIR1 AtFBL1 protein {Arabidopsis thaliana},
partial (39%)
Length = 742
Score = 94.7 bits (234), Expect = 6e-21
Identities = 39/68 (57%), Positives = 51/68 (74%)
Frame = +2
Query: 9 FPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNI 68
FP+EVLE V + S KDR S+SLVCK WY ERW RR VF+GNCY+++P ++ +R +
Sbjct: 206 FPEEVLEHVFSFIDSTKDRGSISLVCKSWYEIERWCRRRVFVGNCYAITPAMVIKRFPKV 385
Query: 69 RSVTLKGK 76
RS+TLKGK
Sbjct: 386 RSITLKGK 409
>TC85267 similar to GP|9279671|dbj|BAB01228.1 transport inhibitor
response-like protein {Arabidopsis thaliana}, partial
(48%)
Length = 1795
Score = 90.5 bits (223), Expect = 1e-19
Identities = 41/68 (60%), Positives = 52/68 (76%)
Frame = +1
Query: 9 FPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNI 68
FPDEV+E V V S DR+S+SLVCK WY ER++R+ VFIGNCYS+SPE L R ++
Sbjct: 925 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 1104
Query: 69 RSVTLKGK 76
+S+TLKGK
Sbjct: 1105KSLTLKGK 1128
>TC80432 similar to PIR|T52139|T52139 LRR-containing F-box protein
[imported] - Arabidopsis thaliana, partial (18%)
Length = 621
Score = 58.2 bits (139), Expect = 6e-10
Identities = 27/66 (40%), Positives = 41/66 (61%)
Frame = +2
Query: 11 DEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNIRS 70
D VL+ V+ + KDR +VS VC+ WY + +R+ V I CY+ +P+ L RR ++ S
Sbjct: 236 DVVLDCVMPYIHDPKDRDAVSQVCRRWYEIDSQTRKHVTIALCYTTTPDRLRRRFPHLES 415
Query: 71 VTLKGK 76
+ LKGK
Sbjct: 416 LKLKGK 433
>TC84158 similar to PIR|T52139|T52139 LRR-containing F-box protein
[imported] - Arabidopsis thaliana, partial (11%)
Length = 614
Score = 42.7 bits (99), Expect = 3e-05
Identities = 18/48 (37%), Positives = 29/48 (59%)
Frame = +3
Query: 11 DEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSP 58
D VL+ V+ + KDR +VS VC+ W+ + +R+ V I CY+ +P
Sbjct: 375 DVVLDCVIPYITDPKDRDAVSQVCRRWHELDSMTRKHVTIALCYTTTP 518
>TC81462 similar to GP|21536497|gb|AAM60829.1 F-box protein family AtFBL4
{Arabidopsis thaliana}, partial (67%)
Length = 1452
Score = 39.3 bits (90), Expect = 3e-04
Identities = 22/64 (34%), Positives = 34/64 (52%), Gaps = 2/64 (3%)
Frame = +2
Query: 10 PDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYS--VSPEILTRRSSN 67
PDE++ + + S+ R + SLVC W ER +R S+ IG S + ++L R N
Sbjct: 116 PDELIVEIFRRLDSKPTRDAASLVCNRWLRLERLTRSSIRIGATGSPDLFVQLLASRFFN 295
Query: 68 IRSV 71
I +V
Sbjct: 296 ITAV 307
>BQ139819
Length = 239
Score = 36.6 bits (83), Expect = 0.002
Identities = 17/53 (32%), Positives = 28/53 (52%)
Frame = +3
Query: 10 PDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILT 62
P+ ++ +L +K DR++VSL CK Y + R S+ +G + E LT
Sbjct: 33 PEHIIWEILNRIKKTSDRNNVSLACKRLYYLDNAQRHSLKVGCGMDPANEALT 191
>TC76362 similar to GP|10716949|gb|AAG21977.1 SKP1 interacting partner 2
{Arabidopsis thaliana}, partial (57%)
Length = 1307
Score = 35.4 bits (80), Expect = 0.004
Identities = 21/69 (30%), Positives = 33/69 (47%), Gaps = 3/69 (4%)
Frame = +1
Query: 1 NILNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFI---GNCYSVS 57
N +N + PDE L +L + + DR S S+VCK W+ E SR+ + + +
Sbjct: 223 NGVNYSEEIPDECLAGILHFLDAG-DRKSCSVVCKRWFRVEGESRQRLSLNAEAKLVDIV 399
Query: 58 PEILTRRSS 66
P + R S
Sbjct: 400 PSLFNRYDS 426
>TC88495 similar to GP|15420162|gb|AAK97303.1 F-box containing protein ORE9
{Arabidopsis thaliana}, partial (12%)
Length = 876
Score = 34.7 bits (78), Expect = 0.007
Identities = 16/48 (33%), Positives = 27/48 (55%)
Frame = +1
Query: 1 NILNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSV 48
N S P+E+L +V + + R+S+SLVC ++ ER +R S+
Sbjct: 175 NSATTVSHLPEEILSKVFTGITDTRTRNSLSLVCHSFFKLERKTRLSL 318
>TC81565 similar to GP|6041850|gb|AAF02159.1| unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 1070
Score = 33.9 bits (76), Expect = 0.012
Identities = 16/63 (25%), Positives = 33/63 (51%), Gaps = 1/63 (1%)
Frame = +3
Query: 10 PDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSV-SPEILTRRSSNI 68
PD+ L + + R DR S L C+ W + ++RR++ +S+ +P L+ + ++
Sbjct: 573 PDDCLTIIFHGLNCRTDRESFGLTCRRWLRIQNFNRRTLQFECSFSILNPSSLSAKGLDV 752
Query: 69 RSV 71
+V
Sbjct: 753 YTV 761
>TC79219 similar to GP|15810000|gb|AAL06927.1 AT5g23340/MKD15_20
{Arabidopsis thaliana}, partial (78%)
Length = 1619
Score = 32.7 bits (73), Expect = 0.027
Identities = 13/38 (34%), Positives = 20/38 (52%)
Frame = +3
Query: 11 DEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSV 48
D+ L +L V S KD+ + LVCK W + R+ +
Sbjct: 147 DDELRSILAKVDSEKDKETFGLVCKRWLRLQSTERKKL 260
>TC89980 similar to PIR|D96512|D96512 hypothetical protein F2G19.16
[imported] - Arabidopsis thaliana, partial (22%)
Length = 980
Score = 32.3 bits (72), Expect = 0.035
Identities = 22/70 (31%), Positives = 33/70 (46%), Gaps = 2/70 (2%)
Frame = +1
Query: 7 SPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEI--LTRR 64
S PDE L V + DR+ SLVC+ W + E SR+ + + + P I L R
Sbjct: 628 SDLPDECLAIVFQSLNP-SDRNQCSLVCRRWLHVEGQSRQRLSLNAKLDLLPVIPSLFNR 804
Query: 65 SSNIRSVTLK 74
++ + LK
Sbjct: 805 FDSVTKLALK 834
>BG646112 weakly similar to GP|10177368|dbj contains similarity to heat
shock transcription factor HSF30~gene_id:K16L22.13,
partial (5%)
Length = 742
Score = 30.0 bits (66), Expect = 0.17
Identities = 17/75 (22%), Positives = 37/75 (48%), Gaps = 4/75 (5%)
Frame = +2
Query: 1 NILNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFI----GNCYSV 56
+++++ S PD +L +L + ++ + SLV + W + W +VF+ NC
Sbjct: 5 DVIDRMSSLPDSLLCHILSFLPTKTSVMTTSLVSRRWRHL--WEHLNVFVFDDKSNCNCR 178
Query: 57 SPEILTRRSSNIRSV 71
+P+ + + + SV
Sbjct: 179NPKKFRKFAFFVSSV 223
>TC82149 weakly similar to PIR|H96695|H96695 hypothetical protein F5A8.10
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1225
Score = 29.6 bits (65), Expect = 0.23
Identities = 13/34 (38%), Positives = 20/34 (58%)
Frame = +3
Query: 10 PDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERW 43
P EV+ +L ++ S +D SL CK+W A R+
Sbjct: 414 PVEVIGNILSLLGSARDVVIASLTCKKWRQAWRY 515
>BI309637 similar to PIR|D86241|D86 protein T16B5.8 [imported] - Arabidopsis
thaliana, partial (28%)
Length = 714
Score = 29.6 bits (65), Expect = 0.23
Identities = 17/59 (28%), Positives = 29/59 (48%)
Frame = +2
Query: 10 PDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNI 68
PD +L+ +L V + +D + + V K W N+ R F N + +P R +S+I
Sbjct: 74 PDVILQYILSHVSNGRDVAYCNCVSKRWKNSMACIRSLYFTRNAFDNAPH---RENSDI 241
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 28.9 bits (63), Expect = 0.38
Identities = 13/47 (27%), Positives = 26/47 (54%)
Frame = +3
Query: 3 LNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVF 49
+++ S PD +L +L + +R +++SL ++W N W + VF
Sbjct: 252 IDRISTLPDPLLLHILSFLPTRTSVATMSLASRKWRNL--WEQLQVF 386
>TC78494
Length = 1450
Score = 28.5 bits (62), Expect = 0.50
Identities = 18/55 (32%), Positives = 25/55 (44%)
Frame = +1
Query: 1 NILNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYS 55
N+ P E++ +L + SR+ + LVCK WYN S F N YS
Sbjct: 163 NVSTSMENLPHELVSNILSRLPSRELLKN-KLVCKTWYNL---ITDSHFTNNYYS 315
>BG581615 similar to GP|10177368|dbj contains similarity to heat shock
transcription factor HSF30~gene_id:K16L22.13, partial
(3%)
Length = 825
Score = 28.5 bits (62), Expect = 0.50
Identities = 13/46 (28%), Positives = 24/46 (51%)
Frame = +3
Query: 4 NQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVF 49
++ S PD +L +L + +R ++++LV + W N W VF
Sbjct: 315 DRLSGLPDSILCHILSFLPTRTSVATMNLVSRRWRNL--WKHAQVF 446
>TC81017 weakly similar to GP|21632795|gb|AAL47185.1
phosphoinositide-dependent protein kinase-1 beta {Mus
musculus}, partial (32%)
Length = 721
Score = 24.6 bits (52), Expect = 7.2
Identities = 10/23 (43%), Positives = 16/23 (69%)
Frame = +2
Query: 44 SRRSVFIGNCYSVSPEILTRRSS 66
SR + F+G VSPE+LT +++
Sbjct: 533 SRAASFVGTAEYVSPELLTSKNA 601
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.131 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,503,605
Number of Sequences: 36976
Number of extensions: 30070
Number of successful extensions: 234
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 234
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 234
length of query: 76
length of database: 9,014,727
effective HSP length: 52
effective length of query: 24
effective length of database: 7,091,975
effective search space: 170207400
effective search space used: 170207400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0104.7


