

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0104.2
(456 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
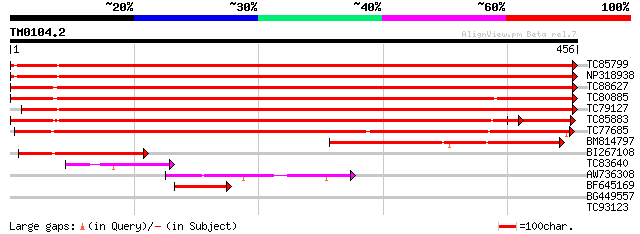
Score E
Sequences producing significant alignments: (bits) Value
TC85799 apyrase-like protein [Medicago truncatula] 659 0.0
NP318938 NP318938|AF288132.1|AAK15160.1 putative apyrase [Medica... 659 0.0
TC88627 apyrase-like protein [Medicago truncatula] 657 0.0
TC80885 apyrase-like protein [Medicago truncatula] 650 0.0
TC79127 apyrase-like protein [Medicago truncatula] 642 0.0
TC85883 apyrase-like protein [Medicago truncatula] 590 e-169
TC77685 apyrase-like protein [Medicago truncatula] 520 e-148
BM814797 similar to GP|10717159|gb apyrase 2 {Pisum sativum}, pa... 230 7e-61
BI267108 homologue to GP|10717159|gb apyrase 2 {Pisum sativum}, ... 99 3e-21
TC83640 similar to GP|14423550|gb|AAK62457.1 putative nucleoside... 59 5e-09
AW736308 similar to GP|19920124|gb| Putative nucleoside phosphat... 54 2e-07
BF645169 similar to GP|10717159|gb| apyrase 2 {Pisum sativum}, p... 50 2e-06
BG449557 similar to GP|19920124|gb| Putative nucleoside phosphat... 31 1.1
TC93123 similar to GP|6728986|gb|AAF26984.1| putative histidyl t... 28 9.6
>TC85799 apyrase-like protein [Medicago truncatula]
Length = 1674
Score = 659 bits (1701), Expect = 0.0
Identities = 323/458 (70%), Positives = 387/458 (83%), Gaps = 2/458 (0%)
Frame = +3
Query: 1 MDFLISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVH 60
M+F+ +L+TF+ +MP+IS SQYLGNNIL+ + + PK QEP++SYAV+FDAGSTGSRVH
Sbjct: 207 MNFM-TLITFLLFIMPSISYSQYLGNNILLTNRKIFPK-QEPISSYAVVFDAGSTGSRVH 380
Query: 61 VYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNT 120
VY+FDQNL+LL V ++EFY+ PGLS+YA NPEEAA+SLIPLL++AE+VVP Q+ T
Sbjct: 381 VYHFDQNLNLLHVGKDVEFYNKTTPGLSAYADNPEEAAKSLIPLLEQAESVVPEDQRSKT 560
Query: 121 PVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINY 180
P++LGATAGLRLL G+A+E ILQ+VRD+ SNRS NVQ DAVSI+DGTQEGSYLWVT+NY
Sbjct: 561 PIRLGATAGLRLLNGDASEKILQSVRDLFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNY 740
Query: 181 LLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDL 240
LGKLGK+FTKTVGV+DLGGGSVQM YAVS+ TAKNAPKV +GEDPYIKKLVL+GKKYDL
Sbjct: 741 ALGKLGKKFTKTVGVMDLGGGSVQMAYAVSKYTAKNAPKVADGEDPYIKKLVLKGKKYDL 920
Query: 241 YVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRK 300
YVHSYL +GREA RAEI KV S NPCILAGFDG YTY+G E+K +APASG+N +C+K
Sbjct: 921 YVHSYLHFGREASRAEILKVTHNSPNPCILAGFDGTYTYAGEEFKANAPASGANFKKCKK 1100
Query: 301 IALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIF-VNKP-NAKI 358
I +ALK+N PCPYQNCTFGGIW+GGGGSGQ+ LF SSF+YL+ED+G+ N P + +
Sbjct: 1101IVREALKLNYPCPYQNCTFGGIWSGGGGSGQRILFAASSFFYLAEDIGLVDPNTPYSLTL 1280
Query: 359 RPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVT 418
RPVDL+T AK AC NLE+AKS YP L + + VEYVC+DL+Y Y LLVDGFGLDP QE+T
Sbjct: 1281RPVDLETEAKKACTLNLEEAKSTYPLLVDFNIVEYVCMDLIYQYVLLVDGFGLDPLQEIT 1460
Query: 419 VANEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 456
+IEYQD+LVEAAWPLG A+EAISSLPKFERLMYFI
Sbjct: 1461AGKQIEYQDSLVEAAWPLGNAVEAISSLPKFERLMYFI 1574
>NP318938 NP318938|AF288132.1|AAK15160.1 putative apyrase [Medicago
truncatula]
Length = 1401
Score = 659 bits (1700), Expect = 0.0
Identities = 321/457 (70%), Positives = 382/457 (83%), Gaps = 1/457 (0%)
Frame = +1
Query: 1 MDFLISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVH 60
M+F+ +L+TF+ +MP+IS SQYLGNNIL+ + + PK QEP++SYAV+FDAGSTGSRVH
Sbjct: 34 MNFM-TLITFLLFIMPSISYSQYLGNNILLTNRKIFPK-QEPISSYAVVFDAGSTGSRVH 207
Query: 61 VYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNT 120
VY+FDQNL+LL V ++EFY+ PGLS+YA NPEEAA+SLIPLL++AE+VVP Q+ T
Sbjct: 208 VYHFDQNLNLLHVGKDVEFYNKTTPGLSAYADNPEEAAKSLIPLLEQAESVVPEDQRSKT 387
Query: 121 PVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINY 180
P++LGATAGLRLL G+A+E ILQ+VRD+ SNRS NVQ DAVSI+DGTQEG YLWVT+NY
Sbjct: 388 PIRLGATAGLRLLNGDASEKILQSVRDLFSNRSTFNVQPDAVSIIDGTQEGCYLWVTVNY 567
Query: 181 LLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDL 240
LG LGK+FTKTVGV+D+GGGSVQM YAVS+ TAKNAPKV +GEDPYIKKLVL+GK YDL
Sbjct: 568 ALGNLGKKFTKTVGVMDVGGGSVQMAYAVSKYTAKNAPKVADGEDPYIKKLVLKGKPYDL 747
Query: 241 YVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRK 300
YVHSYL +GREA RAEI KV S NPC+LAGFDG YTY+G E+K APASG+N N C+K
Sbjct: 748 YVHSYLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKK 927
Query: 301 IALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIFVNK-PNAKIR 359
I KALK+N PCPYQNCTFGGIWNGGGG+GQK+LF +SSF+YL EDVG+ K PN KIR
Sbjct: 928 IIRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIR 1107
Query: 360 PVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTV 419
PVDL + AK AC N EDAKS YP L +K+ YVC+DL+Y Y LLVDGFGLDP QE+T
Sbjct: 1108PVDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGFGLDPLQEITS 1287
Query: 420 ANEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 456
EIEYQDA++EAAWPLG A+EAISSLPKFER+MYF+
Sbjct: 1288GKEIEYQDAVLEAAWPLGNAVEAISSLPKFERMMYFV 1398
>TC88627 apyrase-like protein [Medicago truncatula]
Length = 1614
Score = 657 bits (1694), Expect = 0.0
Identities = 322/458 (70%), Positives = 380/458 (82%), Gaps = 2/458 (0%)
Frame = +1
Query: 1 MDFLISLMTFVFML-MPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRV 59
M+FLI+L+T V +L MPAI+SSQYLGNN+L NRKI + QE ++SYAV+FDAGSTGSR+
Sbjct: 73 MEFLITLITTVLLLLMPAITSSQYLGNNLLTNRKIF--QKQETISSYAVVFDAGSTGSRI 246
Query: 60 HVYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPN 119
HVY+FDQNLDLL + ++EF++ + PGLSSYA +PE+AA+SLIPLL++AENVVP+
Sbjct: 247 HVYHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHK 426
Query: 120 TPVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTIN 179
TP++LGATAGLRLL G+A+E ILQAVRDM SNRS NVQ DAVSI+DGTQEGSYLWVT+N
Sbjct: 427 TPIRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVN 606
Query: 180 YLLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYD 239
Y LG LGK++TKTVGV+DLGGGSVQM YAVS+ TAKNAPKV +G DPYIKKLVL+GK YD
Sbjct: 607 YALGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYD 786
Query: 240 LYVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCR 299
LYVHSYL +GREA RAEI KV S N CIL GFDG YTY+G E+K APASG+N N C+
Sbjct: 787 LYVHSYLHFGREASRAEIMKVTRSSPNTCILTGFDGTYTYAGEEFKAKAPASGANFNGCK 966
Query: 300 KIALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIFVNK-PNAKI 358
KI KALK+N PCPYQNCTFGGIWNGGGG+GQK+L +SSF+YL EDVG+ K PN KI
Sbjct: 967 KIIRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLLASSSFFYLPEDVGMVDPKTPNFKI 1146
Query: 359 RPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVT 418
RPVDL + AK AC N EDAKS YP L +K+ YVC+DL+Y Y LLVDGFGLDP QE+T
Sbjct: 1147RPVDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGFGLDPLQEIT 1326
Query: 419 VANEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 456
EIEYQDA++EAAWPLG A+EAISSLPKFER+MYF+
Sbjct: 1327SGKEIEYQDAVLEAAWPLGNAVEAISSLPKFERMMYFV 1440
>TC80885 apyrase-like protein [Medicago truncatula]
Length = 1451
Score = 650 bits (1676), Expect = 0.0
Identities = 316/458 (68%), Positives = 383/458 (82%), Gaps = 2/458 (0%)
Frame = +2
Query: 1 MDFLISLMT-FVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRV 59
M+FLI+L+T F+ +LMPAI+SSQYLGNN+L NRKI + QE +TSYAVIFDAGSTG+RV
Sbjct: 5 MEFLITLITTFLLLLMPAITSSQYLGNNLLTNRKIF--QKQETLTSYAVIFDAGSTGTRV 178
Query: 60 HVYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPN 119
HVY+FDQNLDLL + N++EF D +KPGLS+YA NPE+AA+SL+PLL+EAE+V+P P
Sbjct: 179 HVYHFDQNLDLLHIGNDIEFVDKIKPGLSAYADNPEQAAKSLLPLLEEAEDVIPEDMHPK 358
Query: 120 TPVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTIN 179
TP++LGATAGLRLL G+AAE ILQA R+M SNRS LNVQSDAVSI+DGTQEGSY+WVT+N
Sbjct: 359 TPLRLGATAGLRLLNGDAAEKILQATRNMFSNRSTLNVQSDAVSIIDGTQEGSYMWVTVN 538
Query: 180 YLLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYD 239
Y+LG LGK FTKTVGV+DLGGGSVQMTYAVS+ TAKNAPKV +GEDPYIKKLVL+GK+YD
Sbjct: 539 YILGNLGKSFTKTVGVIDLGGGSVQMTYAVSKKTAKNAPKVADGEDPYIKKLVLKGKQYD 718
Query: 240 LYVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCR 299
LYVHSYLR+G+EA RA++ GSANPCIL GF+G +TYSG EYK +P+SGSN N+C+
Sbjct: 719 LYVHSYLRFGKEATRAQVLNATNGSANPCILPGFNGTFTYSGVEYKAFSPSSGSNFNECK 898
Query: 300 KIALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVG-IFVNKPNAKI 358
+I LK LKVN PCPY +CTF GIWNGGGGSGQK LF+TS+F YL+EDVG + NKPN+ +
Sbjct: 899 EIILKVLKVNDPCPYSSCTFSGIWNGGGGSGQKKLFVTSAFAYLTEDVGMVEPNKPNSTL 1078
Query: 359 RPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVT 418
P+D + AK AC N ED KS YP L E YVC+DL+Y + LLV GFGL P +E+T
Sbjct: 1079HPIDFEIEAKRACALNFEDVKSTYPRLTEAKR-PYVCMDLLYQHVLLVHGFGLSPRKEIT 1255
Query: 419 VANEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 456
V I+YQ+++VEAAWPLGTA+EAIS+LPKF+RLMYFI
Sbjct: 1256VGEGIQYQNSVVEAAWPLGTAVEAISTLPKFKRLMYFI 1369
>TC79127 apyrase-like protein [Medicago truncatula]
Length = 1501
Score = 642 bits (1655), Expect = 0.0
Identities = 313/448 (69%), Positives = 371/448 (81%), Gaps = 1/448 (0%)
Frame = +3
Query: 10 FVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYNFDQNLD 69
F +MP+IS SQ LGNNIL+ + + PK QE ++SYAV+FDAGSTGSRVHVY+FDQNL+
Sbjct: 81 FYLFIMPSISYSQNLGNNILLTNRKIFPK-QETISSYAVVFDAGSTGSRVHVYHFDQNLN 257
Query: 70 LLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKLGATAG 129
LL V ++EFY+ PGLS+YA NPE+AA+SLIPLL++AE+VVP Q+ TPV+LGATAG
Sbjct: 258 LLHVGKDVEFYNKTTPGLSAYADNPEQAAKSLIPLLEQAESVVPEDQRSKTPVRLGATAG 437
Query: 130 LRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGKLGKRF 189
LRLL G+A+E ILQ+VRD+LSNRS NVQ DAVSI+DGTQEGSYLWVT+NY LG LGK+F
Sbjct: 438 LRLLNGDASEKILQSVRDLLSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGTLGKKF 617
Query: 190 TKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLYVHSYLRYG 249
TKTVGV+DLGGGSVQM YAVSRNTAKNAPKV +G+DPYIKKLVL+GKKYDLYVHSYL +G
Sbjct: 618 TKTVGVMDLGGGSVQMAYAVSRNTAKNAPKVADGDDPYIKKLVLKGKKYDLYVHSYLHFG 797
Query: 250 REAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALKALKVN 309
EA RAEI KV S NPCILAGFDG Y Y+G E+K +A ASG++ +C+KI +ALK+N
Sbjct: 798 TEASRAEILKVTHNSPNPCILAGFDGTYRYAGEEFKANALASGASFKKCKKIVHQALKLN 977
Query: 310 APCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVG-IFVNKPNAKIRPVDLKTAAK 368
PCPYQNCTFGGIWNGGGGSGQ+ LF S F+YL+ +VG + NKPN KIRPVD ++ AK
Sbjct: 978 YPCPYQNCTFGGIWNGGGGSGQRKLFAASFFFYLAAEVGMVDPNKPNFKIRPVDFESEAK 1157
Query: 369 LACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANEIEYQDA 428
AC N EDAKS YP L +K+ YVC+DL+Y Y LLVDGFGLDP QE+T +IEYQD+
Sbjct: 1158KACALNFEDAKSSYPFLAKKNIASYVCMDLIYQYVLLVDGFGLDPLQEITAGKQIEYQDS 1337
Query: 429 LVEAAWPLGTAIEAISSLPKFERLMYFI 456
LVEAAWPLG A+EAISSLPKFE+LMYFI
Sbjct: 1338LVEAAWPLGNAVEAISSLPKFEKLMYFI 1421
>TC85883 apyrase-like protein [Medicago truncatula]
Length = 1848
Score = 590 bits (1522), Expect = e-169
Identities = 295/414 (71%), Positives = 342/414 (82%), Gaps = 1/414 (0%)
Frame = +3
Query: 1 MDFLISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVH 60
M+FLI L+TF+ LMP ISSSQYLGNNIL NRKI PK QE +TSYAV+FDAGSTGSRVH
Sbjct: 246 MEFLIKLITFLLFLMPTISSSQYLGNNILTNRKIF-PK-QETLTSYAVVFDAGSTGSRVH 419
Query: 61 VYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNT 120
VY+FDQNLDLL + N++EFY+ PGLS+YA NP+EAAESLIPLL++AE VVPV+ QP T
Sbjct: 420 VYHFDQNLDLLHIGNDVEFYNKTTPGLSAYADNPKEAAESLIPLLEQAERVVPVNLQPKT 599
Query: 121 PVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINY 180
PVKLGATAGLRLL+GN++E IL+AV +L RS NVQSDAV I+DGTQEGSYLWVTINY
Sbjct: 600 PVKLGATAGLRLLDGNSSELILEAVSSLLKKRSTFNVQSDAVGIIDGTQEGSYLWVTINY 779
Query: 181 LLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDL 240
+LG LGK F++TV V DLGGGSVQM YAVSR AK AP+VP+GEDPYIKK+VL+GKKY L
Sbjct: 780 VLGNLGKDFSETVAVADLGGGSVQMVYAVSREQAKKAPQVPQGEDPYIKKIVLKGKKYYL 959
Query: 241 YVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRK 300
YVHSYLR+G+EA RAEI KV GS NPCILAG+ G YTYSG EYK +PASGSN ++C++
Sbjct: 960 YVHSYLRFGKEASRAEILKVTNGSPNPCILAGYHGTYTYSGEEYKAFSPASGSNFDECKE 1139
Query: 301 IALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIF-VNKPNAKIR 359
I LKALKVN PCPY C+FGGIWNGGGGSGQK L++TSSFYY+ V I NKPN+KIR
Sbjct: 1140IILKALKVNDPCPYGKCSFGGIWNGGGGSGQKTLYVTSSFYYVPTGVNIADPNKPNSKIR 1319
Query: 360 PVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDP 413
DLKT A+ CKT +DAK+ YP +YE DS+ Y CLDL+Y YTL VDGFGLDP
Sbjct: 1320IEDLKTGAEQVCKTKYKDAKATYPLIYE-DSLPYACLDLIYQYTLFVDGFGLDP 1478
Score = 76.3 bits (186), Expect = 2e-14
Identities = 37/55 (67%), Positives = 45/55 (81%)
Frame = +1
Query: 401 VYTLLVDGFGLDPFQEVTVANEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYF 455
++ LL+D P QE+TVAN+IEYQDALV+AAWPLG AIEAISSLPKF+ MY+
Sbjct: 1444 IHCLLMD-LA*TPLQEITVANQIEYQDALVDAAWPLGNAIEAISSLPKFDPFMYY 1605
>TC77685 apyrase-like protein [Medicago truncatula]
Length = 2172
Score = 520 bits (1339), Expect = e-148
Identities = 263/454 (57%), Positives = 348/454 (75%), Gaps = 4/454 (0%)
Frame = +1
Query: 5 ISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYNF 64
I L+TFV +MP+ S+ G+ L+NRK L +++ SYAVIFDAGS+GSRVHV++F
Sbjct: 430 ILLITFVLYMMPSSSNYDSAGDYALLNRK--LSPDKKSGGSYAVIFDAGSSGSRVHVFHF 603
Query: 65 DQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKL 124
DQNLDL+ + +LE ++ +KPGLS+YA P++AAESL+ LL++AE VVP + TPV++
Sbjct: 604 DQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRI 783
Query: 125 GATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGK 184
GATAGLR LEG+A++ IL+AVRD+L +RS+ +DAV++LDGTQEG+Y WVTINYLLG
Sbjct: 784 GATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGN 963
Query: 185 LGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLYVHS 244
LGK ++KTVGVVDLGGGSVQM YA+S + A AP+V +GEDPY+K++ L+G+KY LYVHS
Sbjct: 964 LGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHS 1143
Query: 245 YLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALK 304
YLRYG A RAEI KVAG + NPCIL+G DG Y Y G +K S +SG++LN+C+ +A K
Sbjct: 1144YLRYGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKAS--SSGASLNECKSVAHK 1317
Query: 305 ALKVN-APCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVG-IFVNKPNAKIRPVD 362
ALKVN + C + CTFGGIWNGGGG GQKNLF+ S F+ + + G + N P A +RP D
Sbjct: 1318ALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPAD 1497
Query: 363 LKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANE 422
+ AAK AC+T LE+AKS YP + E+ ++ Y+C+DLVY YTLLVDGFG+ P+QE+T+ +
Sbjct: 1498FEDAAKQACQTKLENAKSTYPRV-EEGNLPYLCMDLVYQYTLLVDGFGIYPWQEITLVKK 1674
Query: 423 IEYQDALVEAAWPLGTAIEAISSL--PKFERLMY 454
++Y DALVEAAWPLG+AIEA+SS +F L Y
Sbjct: 1675VKYDDALVEAAWPLGSAIEAVSST*QDRFHELGY 1776
>BM814797 similar to GP|10717159|gb apyrase 2 {Pisum sativum}, partial (42%)
Length = 690
Score = 230 bits (587), Expect = 7e-61
Identities = 112/192 (58%), Positives = 142/192 (73%), Gaps = 3/192 (1%)
Frame = +1
Query: 258 FKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALKALKVN-APCPYQN 316
FK + NPCILAG+DG+Y Y G + S+ SGS+LN+C+ IAL AL VN + C +
Sbjct: 1 FKASDDFGNPCILAGYDGSYKYGGKSFNASSSPSGSSLNECKSIALNALTVNESTCAHMK 180
Query: 317 CTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIFVN--KPNAKIRPVDLKTAAKLACKTN 374
CTFGGIWNGGGG GQKNLF+ S F+ + G F N P AK+RPVD K AAK AC+T
Sbjct: 181 CTFGGIWNGGGGDGQKNLFVASFFFDRAAQAG-FANPKSPVAKVRPVDFKNAAKQACQTK 357
Query: 375 LEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANEIEYQDALVEAAW 434
LEDAKS YP L + ++ Y+C+DLVY YTLLVDGFGLDP Q++T+ +++Y D+LVEAAW
Sbjct: 358 LEDAKSTYP-LVDDGNLPYLCMDLVYQYTLLVDGFGLDPLQDITLVKQVKYHDSLVEAAW 534
Query: 435 PLGTAIEAISSL 446
PLG+AIEA+SS+
Sbjct: 535 PLGSAIEAVSSI 570
>BI267108 homologue to GP|10717159|gb apyrase 2 {Pisum sativum}, partial
(24%)
Length = 449
Score = 99.4 bits (246), Expect = 3e-21
Identities = 48/104 (46%), Positives = 79/104 (75%)
Frame = +2
Query: 8 MTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYNFDQN 67
+T + +MP+ SS++ + + L +RKI +++ S+AV+FDAGS+GSRVHV+ FD+N
Sbjct: 128 VTLILYIMPSTSSNESIEDYALTHRKI--SPDRKISDSFAVVFDAGSSGSRVHVFRFDRN 301
Query: 68 LDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENV 111
L+L+ + N+LE + +KPGLS+YA +P++AA+SL+ LL +AE+V
Sbjct: 302 LELVKIGNDLEVFLQIKPGLSAYARDPQQAAKSLVSLLDKAESV 433
>TC83640 similar to GP|14423550|gb|AAK62457.1 putative nucleoside
triphosphatase {Arabidopsis thaliana}, partial (12%)
Length = 601
Score = 58.5 bits (140), Expect = 5e-09
Identities = 33/93 (35%), Positives = 52/93 (55%), Gaps = 6/93 (6%)
Frame = +3
Query: 46 YAVIFDAGSTGSRVHVYNFDQNLDLLPVENELEFYDS------VKPGLSSYAANPEEAAE 99
Y +I D GSTG+RVHV+ + ++N L+F V PGLSS+ +P+ A
Sbjct: 342 YRIIIDGGSTGTRVHVFKYK-------MKNALDFGKKGLVSMRVNPGLSSFGNDPDGAGR 500
Query: 100 SLIPLLKEAENVVPVSQQPNTPVKLGATAGLRL 132
SL+ ++ A+ +P T ++L ATAG+R+
Sbjct: 501 SLLEVVDFAKRRIPKENWRETEIRLMATAGMRM 599
>AW736308 similar to GP|19920124|gb| Putative nucleoside phosphatase {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (3%)
Length = 552
Score = 53.5 bits (127), Expect = 2e-07
Identities = 43/168 (25%), Positives = 76/168 (44%), Gaps = 15/168 (8%)
Frame = +1
Query: 126 ATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGKL 185
ATAG+R + + +L+ V + + S + + +L G +E Y WV +NY +G+
Sbjct: 7 ATAGMRRIHRDDVFRVLEDVEAVAKDHSFM-FDKRWIRVLSGREEAYYGWVALNYNMGRF 183
Query: 186 G---KRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLYV 242
R + T+G+VDLGG S+Q+ + R+T G+D + ++ +
Sbjct: 184 DDDYHRGSSTLGLVDLGGSSLQIVVEIDRDT---------GDDVNAIRSEFGSIEHRIVA 336
Query: 243 HSYLRYG-REAF-----------RAEIFKVAGGSANPCILAGFDGAYT 278
+S +G EAF R E + +PC+++ F YT
Sbjct: 337 YSLPSFGLNEAFDRTVAMLRNNQRVESTRGVSELRHPCLMSTFVQNYT 480
>BF645169 similar to GP|10717159|gb| apyrase 2 {Pisum sativum}, partial (10%)
Length = 542
Score = 49.7 bits (117), Expect = 2e-06
Identities = 22/46 (47%), Positives = 32/46 (68%)
Frame = +1
Query: 133 LEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTI 178
LEG+ ++ ILQAV D+L RS L + + V + DGTQEG++ WV +
Sbjct: 235 LEGDVSDRILQAV*DLLKQRSTLKSEPNVVDVQDGTQEGAFQWVCL 372
>BG449557 similar to GP|19920124|gb| Putative nucleoside phosphatase {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (4%)
Length = 662
Score = 30.8 bits (68), Expect = 1.1
Identities = 12/18 (66%), Positives = 15/18 (82%)
Frame = +1
Query: 46 YAVIFDAGSTGSRVHVYN 63
Y V+ D GSTG+RV+VYN
Sbjct: 469 YYVVLDCGSTGTRVYVYN 522
>TC93123 similar to GP|6728986|gb|AAF26984.1| putative histidyl tRNA
synthetase {Arabidopsis thaliana}, partial (31%)
Length = 644
Score = 27.7 bits (60), Expect = 9.6
Identities = 12/53 (22%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Frame = +1
Query: 209 VSRNTAKNAPKVPEGEDPYIK-KLVLQGKKYDLYVHSYLRYGREAFRAEIFKV 260
V N ++ PK+P+G + K ++ ++ K + + + R+G A +F++
Sbjct: 217 VESNESRRLPKIPKGTRDFAKEQMTIRKKAFSVIEEVFERHGATALDTPVFEL 375
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,394,378
Number of Sequences: 36976
Number of extensions: 158493
Number of successful extensions: 871
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 840
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 847
length of query: 456
length of database: 9,014,727
effective HSP length: 99
effective length of query: 357
effective length of database: 5,354,103
effective search space: 1911414771
effective search space used: 1911414771
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0104.2


