

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
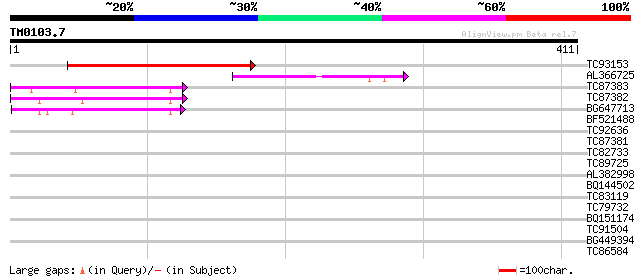
Query= TM0103.7
(411 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 141 4e-34
AL366725 75 5e-14
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 50 1e-06
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 50 2e-06
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 46 2e-05
BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila m... 37 0.018
TC92636 homologue to GP|15042313|gb|AAK82093.1 232R {Chilo iride... 34 0.091
TC87381 32 0.59
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 30 1.7
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 30 2.2
AL382998 similar to GP|23491723|db formin homology protein A {Di... 29 2.9
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 29 3.8
TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 28 5.0
TC79732 weakly similar to GP|6729021|gb|AAF27017.1| hypothetical... 28 5.0
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 28 6.5
TC91504 similar to GP|21618032|gb|AAM67082.1 unknown {Arabidopsi... 28 6.5
BG449394 similar to PIR|A83006|A830 hypothetical protein PA5121 ... 28 6.5
TC86584 similar to GP|11994215|dbj|BAB01337. contains similarity... 28 8.5
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 141 bits (356), Expect = 4e-34
Identities = 64/136 (47%), Positives = 90/136 (66%)
Frame = +2
Query: 43 LDKFLKRSPPKFEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTS 102
L+ FL+ PP F+G Y PDGA +W++E+ERIF + C E +KV F +++L+ EA WW S
Sbjct: 107 LETFLRNHPPTFKGRYAPDGA*KWLKEIERIFRVMQCFETQKVQFGTHMLAEEADDWWIS 286
Query: 103 VRGRITPEEGELIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQ 162
+ + ++ + W +F+ FL +YFP D + +KE+EF ELKQ MSV EYAAKF EL
Sbjct: 287 LLPVLEQDDAVVTWAMFRKEFLGRYFPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELAT 466
Query: 163 FHPRYSTANDETSKCV 178
F+P YS E SKC+
Sbjct: 467 FYPHYSAETAEFSKCI 514
>AL366725
Length = 485
Score = 75.1 bits (183), Expect = 5e-14
Identities = 44/140 (31%), Positives = 66/140 (46%), Gaps = 12/140 (8%)
Frame = +2
Query: 162 QFHPRYSTANDETSKCVKFEYGLRPDIRTVVGHDQIRNFATLVKKCRIFEENEKT*KEYL 221
+F+P Y+ E SKC+KFE GLRPDI+ +G+ Q+R F LV CRI+EE+ K + +
Sbjct: 2 KFYPHYAAETAEFSKCIKFENGLRPDIKRAIGYQQLRVFPDLVNTCRIYEEDTKAHDKVV 181
Query: 222 KGLGVNKAPKKKEEKRKPYF*PQDQGRNQFGRTWNPTG----------GFGFQGRPGGF- 270
+ K + + KPY P D+G+ + P +G +G
Sbjct: 182 N----ERKTKGQ*SRPKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNYGEKGHKSNVC 349
Query: 271 -NNTPFCGRCRRNGHRAQEC 289
C RC + GH +C
Sbjct: 350 PKEIKKCVRCDKKGHIVADC 409
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 50.4 bits (119), Expect = 1e-06
Identities = 38/142 (26%), Positives = 58/142 (40%), Gaps = 13/142 (9%)
Frame = -2
Query: 1 EEMAAMARALEQLT-------AFLTEQAERARQGAGQGAANNQEEIYHGLDKF----LKR 49
+EM M R ++QL A L Q +R + + + +F +K
Sbjct: 910 QEMEKMRR*IQQLQEIVNAQQALLEAQQKRFKDHVSSSDSLSSRSSRSQRREFQMNDIK* 731
Query: 50 SPPKFEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITP 109
P FEG PD +W++ +ER+F+ E +KV + L A WW +V+ R
Sbjct: 730 DIPDFEGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRRRKR 551
Query: 110 EEGELI--WEIFKNSFLEKYFP 129
E I WE + KY P
Sbjct: 550 EGKSKIKTWEKMRQKLTRKYLP 485
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 50.1 bits (118), Expect = 2e-06
Identities = 37/142 (26%), Positives = 61/142 (42%), Gaps = 13/142 (9%)
Frame = +2
Query: 1 EEMAAMARALEQLTAFLTEQ-----AERAR-QGAGQGAANNQEEIYHGLDKFLKRSP--- 51
+EM M R ++QL + Q AE+ R +G + ++ H + L+ +
Sbjct: 443 QEMEDMRRQIQQLQEIINAQQALLEAEQRRFEGDVSYSDSSSSRSSHSQRRQLQMNDIKV 622
Query: 52 --PKFEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITP 109
P FEG D +W++ +ER+FE E +KV + L A WW +++ R
Sbjct: 623 DIPDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRRRKR 802
Query: 110 EEGELI--WEIFKNSFLEKYFP 129
E I W+ + KY P
Sbjct: 803 EGKSKIKTWDKMRQKLTRKYLP 868
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 46.2 bits (108), Expect = 2e-05
Identities = 35/139 (25%), Positives = 60/139 (42%), Gaps = 13/139 (9%)
Frame = +2
Query: 2 EMAAMARALEQLTAFLTEQ-----AERARQ---GAGQGAANNQEEIYHGLD---KFLKRS 50
EM M R ++ L + Q A+R R G+G +++++ H +K
Sbjct: 164 EMEEMRRQIQLLQETVNAQQALLEAQRRRNDDDGSGSDSSSSRSSRSHRRQTRMSKIKVD 343
Query: 51 PPKFEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPE 110
P F G PD +W++ +ER+F+ AE +KV + L A WW +++ + E
Sbjct: 344 IPDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKVKIVAAKLKKHASIWWKNLKRKRNCE 523
Query: 111 EGELI--WEIFKNSFLEKY 127
I W+ + KY
Sbjct: 524 GKSKIKTWDKMRQKLTRKY 580
>BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila
melanogaster}, partial (84%)
Length = 470
Score = 36.6 bits (83), Expect = 0.018
Identities = 29/71 (40%), Positives = 31/71 (42%), Gaps = 1/71 (1%)
Frame = +2
Query: 247 GRNQFGRTWNPTGGFGFQGRP-GGFNNTPFCGRCRRNGHRAQECIVVLGGQSGVQTGGSG 305
GR FG +NP GGFG GRP GGF F GR R G GG G S
Sbjct: 194 GRPGFGGGYNPYGGFG-GGRPYGGFGGGGFGGRPFRGG----------GGGGGGSASASA 340
Query: 306 NPNPTTTVGGG 316
+ N GGG
Sbjct: 341 SAN---AAGGG 364
>TC92636 homologue to GP|15042313|gb|AAK82093.1 232R {Chilo iridescent
virus}, partial (1%)
Length = 772
Score = 34.3 bits (77), Expect = 0.091
Identities = 24/84 (28%), Positives = 39/84 (45%), Gaps = 10/84 (11%)
Frame = +3
Query: 2 EMAAMARALEQLTAFLTEQ-----AERAR---QGAGQGAANNQEEIYHG--LDKFLKRSP 51
EM M R +++L + Q AER R G+ +++ + L +K
Sbjct: 258 EMEEMRRQIQELQETVNAQQAILEAERRRVDDDGSSDSSSSRSSRSHRRKTLMNDIKVDI 437
Query: 52 PKFEGGYNPDGAYEWVRELERIFE 75
P FEG PD +W++ +ER+FE
Sbjct: 438 PDFEGELQPDEFVDWLQAIERVFE 509
>TC87381
Length = 814
Score = 31.6 bits (70), Expect = 0.59
Identities = 17/60 (28%), Positives = 27/60 (44%)
Frame = +3
Query: 47 LKRSPPKFEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGR 106
+K P FEG D + ++ +E +FE E KV + L A WW +++ R
Sbjct: 621 IKVDIPDFEGELQSDEFVD*LQAIECVFEYKEIPEDHKVKVVAV*LKKHALIWWENLKRR 800
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 30.0 bits (66), Expect = 1.7
Identities = 11/14 (78%), Positives = 11/14 (78%)
Frame = +3
Query: 276 CGRCRRNGHRAQEC 289
C RCRR GHRAQ C
Sbjct: 357 CLRCRRRGHRAQNC 398
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 29.6 bits (65), Expect = 2.2
Identities = 17/42 (40%), Positives = 19/42 (44%)
Frame = +1
Query: 276 CGRCRRNGHRAQECIVVLGGQSGVQTGGSGNPNPTTTVGGGS 317
C C +GH A+EC GG G GG G GGGS
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGG--GGGS 135
>AL382998 similar to GP|23491723|db formin homology protein A {Dictyostelium
discoideum}, partial (3%)
Length = 355
Score = 29.3 bits (64), Expect = 2.9
Identities = 17/51 (33%), Positives = 20/51 (38%), Gaps = 6/51 (11%)
Frame = +3
Query: 294 GGQSGVQTGGSGNPNPTTTVGGG------SNRNANVPPRDNRRVGHPANKG 338
GG+ Q G G P P GGG NR PP R+ P +G
Sbjct: 18 GGEGRKQRKGGGGPPPPPGGGGGPPPPPKKNRGGGPPPGVPRKFSPPPKRG 170
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 28.9 bits (63), Expect = 3.8
Identities = 20/53 (37%), Positives = 21/53 (38%)
Frame = +1
Query: 253 RTWNPTGGFGFQGRPGGFNNTPFCGRCRRNGHRAQECIVVLGGQSGVQTGGSG 305
R W P G GR G P G G A +C LGG G TGG G
Sbjct: 409 RGW-PAGSGALGGRRGARGRAPRGGG--GGGGAASDCGGALGGDGGGGTGGGG 558
>TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 421
Score = 28.5 bits (62), Expect = 5.0
Identities = 10/21 (47%), Positives = 13/21 (61%)
Frame = +3
Query: 269 GFNNTPFCGRCRRNGHRAQEC 289
GF+ C C+R GH A+EC
Sbjct: 327 GFSRDNLCKNCKRPGHYAREC 389
>TC79732 weakly similar to GP|6729021|gb|AAF27017.1| hypothetical protein
{Arabidopsis thaliana}, partial (19%)
Length = 654
Score = 28.5 bits (62), Expect = 5.0
Identities = 12/38 (31%), Positives = 24/38 (62%)
Frame = +1
Query: 106 RITPEEGELIWEIFKNSFLEKYFPADAKCRKEMEFFEL 143
++ P +GE W +FKN ++ Y A++ + ++EF E+
Sbjct: 328 KVYPRKGET-WALFKNWDIKWYMDAESHQKYDLEFVEI 438
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 28.1 bits (61), Expect = 6.5
Identities = 16/49 (32%), Positives = 22/49 (44%), Gaps = 5/49 (10%)
Frame = +1
Query: 295 GQSGVQTGGSGNPNPTTTVGGGSNRNANVPP-----RDNRRVGHPANKG 338
G++ ++ G P PT GGG R A PP R+ + G P G
Sbjct: 568 GETHKKSPGGRTPPPTPRPGGGGARGAPAPPPQKKQREGGKSGPPRAPG 714
>TC91504 similar to GP|21618032|gb|AAM67082.1 unknown {Arabidopsis
thaliana}, partial (37%)
Length = 863
Score = 28.1 bits (61), Expect = 6.5
Identities = 13/34 (38%), Positives = 18/34 (52%), Gaps = 1/34 (2%)
Frame = +3
Query: 95 EARTW-WTSVRGRITPEEGELIWEIFKNSFLEKY 127
E ++W WT V G I P L+W + F+E Y
Sbjct: 546 EIKSWGWTEVYGYIMPFVWMLLWSLVLKFFIEVY 647
>BG449394 similar to PIR|A83006|A830 hypothetical protein PA5121 [imported] -
Pseudomonas aeruginosa (strain PAO1), partial (1%)
Length = 684
Score = 28.1 bits (61), Expect = 6.5
Identities = 14/33 (42%), Positives = 18/33 (54%), Gaps = 2/33 (6%)
Frame = -2
Query: 256 NPTGGFGFQGRPGGFNNTPFCGRC--RRNGHRA 286
+P G +G P F+ PFCGR RR G R+
Sbjct: 512 SPNSRRGGRGAPSLFSTAPFCGRSSRRRGGGRS 414
>TC86584 similar to GP|11994215|dbj|BAB01337. contains similarity to
RNA-binding protein~gene_id:MOA2.5 {Arabidopsis
thaliana}, partial (72%)
Length = 1574
Score = 27.7 bits (60), Expect = 8.5
Identities = 12/25 (48%), Positives = 14/25 (56%)
Frame = +2
Query: 265 GRPGGFNNTPFCGRCRRNGHRAQEC 289
G PG FN+ FCG C NG + C
Sbjct: 1115 GGPGAFNSNCFCGIC--NGRKCHCC 1183
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,243,230
Number of Sequences: 36976
Number of extensions: 177919
Number of successful extensions: 868
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 842
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 865
length of query: 411
length of database: 9,014,727
effective HSP length: 98
effective length of query: 313
effective length of database: 5,391,079
effective search space: 1687407727
effective search space used: 1687407727
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0103.7


