

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
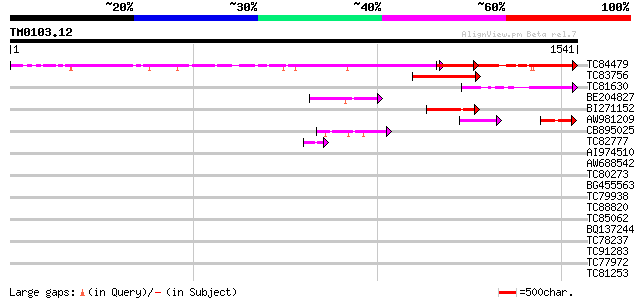
Query= TM0103.12
(1541 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84479 ENBP1 973 0.0
TC83756 similar to PIR|D86254|D86254 hypothetical protein [impor... 224 2e-58
TC81630 similar to PIR|T06461|T06461 DNA-binding protein PD3 ch... 218 2e-56
BE204827 similar to PIR|F86222|F86 hypothetical protein [importe... 159 1e-38
BI271152 weakly similar to PIR|T05151|T051 hypothetical protein ... 134 3e-31
AW981209 similar to PIR|F86222|F86 hypothetical protein [importe... 120 5e-27
CB895025 similar to PIR|F86222|F862 hypothetical protein [import... 102 1e-21
TC82777 weakly similar to PIR|D85438|D85438 hypothetical protein... 44 6e-04
AI974510 42 0.001
AW688542 similar to PIR|T05151|T051 hypothetical protein F18E5.5... 37 0.077
TC80273 similar to GP|8978267|dbj|BAA98158.1 contains similarity... 29 0.18
BG455563 similar to PIR|T05151|T051 hypothetical protein F18E5.5... 35 0.29
TC79938 similar to GP|17933299|gb|AAL48232.1 AT4g17800/dl4935c {... 34 0.38
TC88820 homologue to PIR|D84890|D84890 probable AT-hook DNA-bind... 33 0.85
TC85062 similar to EGAD|119543|127780 hypothetical protein F49C5... 33 0.85
BQ137244 similar to GP|20161222|dbj Epstein-Barr virus EBNA-1-li... 33 0.85
TC78237 similar to GP|17933299|gb|AAL48232.1 AT4g17800/dl4935c {... 33 0.85
TC91283 weakly similar to GP|21554159|gb|AAM63238.1 unknown {Ara... 33 1.1
TC77972 similar to GP|6175162|gb|AAF04888.1| hypothetical protei... 33 1.1
TC81253 similar to GP|9757941|dbj|BAB08429.1 gene_id:MJC20.6~unk... 32 2.5
>TC84479 ENBP1
Length = 5108
Score = 973 bits (2515), Expect = 0.0
Identities = 620/1330 (46%), Positives = 767/1330 (57%), Gaps = 150/1330 (11%)
Frame = +1
Query: 1 MDETGE-ECRRCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRS 59
MDETG+ E RRC R GWRC E+AL GK+ CERH Y +K S ++
Sbjct: 1 MDETGDDESRRCSRNGSGGWRCKEQALPGKTHCERHHEY-------YKSRNSSSFVEKNG 159
Query: 60 GRRKPVDNSENGVV----DDGCKELFG-DPNGTPTVVDEFTGLCGVSEGDAGVNLNLGCE 114
G R NGVV D+G K LFG D +G VV+ F G+ G E + GVN +G E
Sbjct: 160 GIR-------NGVVVDDHDNGGKGLFGGDDHG---VVEGFGGVFGDVEVNGGVNAGIGRE 309
Query: 115 SLNLQDKGEEGQQVHSGGFGEGCGRMGQVLGDYGVEYA----EDRNAVAGLG-------A 163
NL +G+ GQQ G F + G +GQ LGD GVE+ EDRN GLG
Sbjct: 310 RFNLWQQGD-GQQ--GGRFEQASGNLGQFLGD-GVEFVGGFVEDRNRAVGLGQQWGGVGV 477
Query: 164 FRN------VGNEDHGCVAGRNVCVNDRLGLPSEGIESLIGEEPGFG-----SFQALLCK 212
F N VG +DHG VC ND G S+GI LIGE FG SFQALL +
Sbjct: 478 FGNGGGVSGVGKDDHGNGVD-GVCGNDSPGFGSDGINGLIGEGGCFGNLYDRSFQALLSQ 654
Query: 213 DRGCAEDVIFIGDVTGFEGLSGENTHGFRDEVGGFVENPCFEGEND--SNKEGPGSNYKM 270
R C EDV G T F+GL GE+ + FR VG + FEGE + S P S+ KM
Sbjct: 655 GRVCDEDVNLTGGGTSFQGLGGESAYDFRG-VGNLSQCGKFEGEKNVGSILTVPESSNKM 831
Query: 271 SALGFEEEIGLLLSRGGTTNEEARCEALRPLSKRGRPKGSKNENDNKQLSTALDGQSVGG 330
A G EE + +LLS G + NEEAR EAL+PL+KRGRPKGSKN+ K++ +G++V G
Sbjct: 832 GAFGVEEGMEMLLSGGVSINEEARGEALKPLAKRGRPKGSKNKIKKKEVDLVTNGETVCG 1011
Query: 331 DDNAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAEKSGRP------------ 378
N GT +V L E +V SG+ D+ E D+G+ AR +K G+P
Sbjct: 1012 SANVGT----TVEILETEKSVFSGKADQ---EGVDMGDIARTKKRGQPEDWKRGRHIILA 1170
Query: 379 ---------------------KVSKNKIRRVEHVGNVV--AVKIVGPKKHGRPKSSKCRK 415
K S N+ + VE V + V A +I PKK GR ++SKC K
Sbjct: 1171 VGYEIDGVGEITGPMERGRKSKGSVNEEKNVEEVSSEVAGAGEIARPKKLGRMEASKCGK 1350
Query: 416 KNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNN---------EGSGAGEIVRPKK 466
+ ++E ++ GEI KK GRPKGS + ++ NN E +GAGEI RPKK
Sbjct: 1351 EIVVEVSNDVGGEIVRRKKRGRPKGSKCGKEIVLEVNNEVVGAEVIIEVAGAGEIARPKK 1530
Query: 467 RGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSAD 526
GR GSK ++EVS A +I PKK GRPKGSK ++ +V+V+ EVAG+
Sbjct: 1531 LGRLEGSKCGKEIVVEVSNDVAG----EIVRPKKRGRPKGSKCGKEIVVEVNHEVAGAG- 1695
Query: 527 CEIAGPKKCGRPKGSMKKRKSLVCASILEGAGGITREGLENKML--SNLCQEHIEYTQPV 584
EIA KK GRPKG +++ ++ +G ++ LE++ L L Q+ ++ +P
Sbjct: 1696 -EIARSKKRGRPKGYKCQKEIVIKRGRPKGTKN-KKKILEDQELHVQTLVQDEVQNVKPK 1869
Query: 585 VRGGRPKGSRNKKIKLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIG-----KPIG 639
+ GRPKGS+NKK +A +D NK + +E+K G G K I
Sbjct: 1870 L--GRPKGSKNKKKNIAGED-------GNK--------LHKEKKRRGWPKGFCLKPKEIA 1998
Query: 640 LDNDKATLASDRDQET---PNQTLAQDEVQNDKSSVKPKRGRPKGSKNKMKSIANKARNK 696
D+ R + + P +T Q + + + +RGRPKG+ K K I + K
Sbjct: 1999 ARLDEKIERRGRPKGSGMKPKETAVQLDAKIE------RRGRPKGAGKKPKEIVVRLDTK 2160
Query: 697 FGKVRNMRGRPKGSLRKKNETAYCLDSQNERNS---LDGRTSTEAAYRN----------- 742
+ RGRPKGS +K+ E A L Q E +DG ST +++
Sbjct: 2161 IER----RGRPKGSGKKQKEVASQLALQIESQKSTRVDGALSTIVPHKHIQEESISPLKD 2328
Query: 743 ------------------------------DVDLHRGHCSQEELLRMLSVEHKNIQGVGV 772
D+H+ CS E LR L +HKN Q V V
Sbjct: 2329 PVNKEEKSDFVLECSKDSGIEKITKGLMSKSGDVHK-RCS--ERLRTLLTDHKNSQDVEV 2499
Query: 773 EET---------IDYGLRSSGLMGDTERKKETRILRCHQCWRNSWSGVVICAKCKRKQYC 823
EET ID+ L SS LMG+ E KKE R LRCHQCW+ S +G+V+C KCKRK+YC
Sbjct: 2500 EETFCENEVEEAIDHELESSDLMGEPETKKEPRNLRCHQCWKKSRTGIVVCTKCKRKKYC 2679
Query: 824 YECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKDISVMTGSGEADTGVILQKLLYLLN 883
YECI KWY KTREEIE ACPFCL CNCRLCLKK IS M G+GEAD V LQKL YLL
Sbjct: 2680 YECIAKWYQDKTREEIETACPFCLDYCNCRLCLKKTISTMNGNGEADADVKLQKLFYLLK 2859
Query: 884 KTLPLLQHIQREQISEMEVEASMHGSPLMEED------------IQFDNCNTSIVNFHRS 931
KTLPLLQHIQREQ SE+EVEAS+HGS ++EE + DNCNTSIVNFHRS
Sbjct: 2860 KTLPLLQHIQREQKSELEVEASIHGSLMVEEKDILQAAVDDDDRVYCDNCNTSIVNFHRS 3039
Query: 932 CPNPNCRYDLCLTCCMELRNGLHYEDIPAS-GNEETIDEPPITSAWRAEINGRIPCPPKA 990
C NP CRYDLCLTCC ELRNG+H +DIPAS GNEE ++ PP T AWRAE NG IPCPPKA
Sbjct: 3040 CVNPYCRYDLCLTCCTELRNGVHSKDIPASGGNEEMVNTPPETIAWRAETNGSIPCPPKA 3219
Query: 991 RGGCGTSILSLRRLFEANWVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSV 1050
RGGCGT+ LSLRRLF+ANW+ KL R+AEELTI+Y PP VDL + C +C F D A NS
Sbjct: 3220 RGGCGTATLSLRRLFKANWIEKLTRDAEELTIKYQPPIVDLSLECSECRSFEEDAAHNSA 3399
Query: 1051 RKAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMW 1110
RKAASRET HDN LYCPDA+++GDTE++HFQRHWIRGEPVIVRNV++K SGLSW PMVMW
Sbjct: 3400 RKAASRETGHDNLLYCPDAIEIGDTEFDHFQRHWIRGEPVIVRNVYKKGSGLSWDPMVMW 3579
Query: 1111 RAFRGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPP 1170
RAFR A ILK+E TFKAIDCLDWCEVQ+N FQFFKGYL GRRYRNGWP ++ K
Sbjct: 3580 RAFRLAKNILKDEADTFKAIDCLDWCEVQVNAFQFFKGYLTGRRYRNGWPGNVEAKGLAS 3759
Query: 1171 SNSFEECFPR 1180
+ F F +
Sbjct: 3760 NKFFRRLFAK 3789
Score = 296 bits (757), Expect = 5e-80
Identities = 163/311 (52%), Positives = 194/311 (61%), Gaps = 32/311 (10%)
Frame = +3
Query: 1263 PKIIKKLQKKYEAEDMRDLYGRINKTVVSHRSKHKKCRTGISMDPKIPENDDTMGRNSNL 1322
P+IIKKL+KKYE EDMR+LYG +K S K KK R +++D KI E +D GR+S L
Sbjct: 4038 PRIIKKLKKKYEVEDMRELYGLDSKAAGSRGRKRKKRRVRVTVDLKISEKEDINGRDSTL 4217
Query: 1323 RGSQSNEEIVVNELSTRSSSLGESRSDSAACVQGFSESSESKSVLNAGEQAILNMYKRFV 1382
SQ E+ + D ACVQ FSES++SK LN Q +++ RF
Sbjct: 4218 LESQEKED----------------KLDREACVQEFSESTKSKLDLNVSNQEVIDS-PRFQ 4346
Query: 1383 KFDLNNHDSGYLFPGKDCEWMHYDVNNGKQWCSSP-------------VMPC-------- 1421
+FDLN+ DS +L P DCE M YD N +Q CS P PC
Sbjct: 4347 QFDLNSLDSNFLVPRNDCESMLYD--NVEQRCSRPRDGSCKGNTSVIDNQPCGGTKETTF 4520
Query: 1422 -----------SKIQIDKTVPVKNDISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTE 1470
S I+ DK V+N++ SNN ND H+ETQ+GSAVWDIFRRQDVPKLTE
Sbjct: 4521 VNGLDSSDISSSDIETDKIESVENEMPSNNLCGNDVHLETQYGSAVWDIFRRQDVPKLTE 4700
Query: 1471 YLKKHHKEFRHANNLPVDSVTHPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFI 1530
YL KHH+EFRH +LPV+ V HPIHDQ YLNEKHK+QLK EYG+EPWTFEQHLGEAVFI
Sbjct: 4701 YLNKHHREFRHITSLPVNFVIHPIHDQHFYLNEKHKKQLKLEYGVEPWTFEQHLGEAVFI 4880
Query: 1531 PAGCPHQVRNR 1541
PAGCPHQVRNR
Sbjct: 4881 PAGCPHQVRNR 4913
Score = 197 bits (501), Expect = 3e-50
Identities = 92/112 (82%), Positives = 100/112 (89%)
Frame = +2
Query: 1161 EMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPAVLKPDLGPKT 1220
EMLKLKDWPP+N FE+C PRHGAEF MLPFSDYTHPK GILNLATKLP VLKPDLGPKT
Sbjct: 3731 EMLKLKDWPPTNFFEDCLPRHGAEFTTMLPFSDYTHPKSGILNLATKLPTVLKPDLGPKT 3910
Query: 1221 YIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKK 1272
YIAYG+LEELSRGDSVTKLHCDISDAVNILTHT +VK P WQ K KK++++
Sbjct: 3911 YIAYGALEELSRGDSVTKLHCDISDAVNILTHTADVKTPAWQSKNHKKVKEE 4066
>TC83756 similar to PIR|D86254|D86254 hypothetical protein [imported] -
Arabidopsis thaliana, partial (21%)
Length = 775
Score = 224 bits (572), Expect = 2e-58
Identities = 114/188 (60%), Positives = 131/188 (69%), Gaps = 2/188 (1%)
Frame = +1
Query: 1095 VFEKASGLSWHPMVMWRAF-RGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGR 1153
V + +GLSW PMVMWRA + + + KAIDC+ CEV IN FFKGY+EGR
Sbjct: 1 VLKHGTGLSWEPMVMWRALCDNLASDISSKMSEVKAIDCMANCEVAINTRMFFKGYIEGR 180
Query: 1154 RYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPA-VL 1212
Y N WPEMLKLKDWPPS+ FE+ PRH EFI LPF YT P+ G LNLA KLPA VL
Sbjct: 181 TYGNLWPEMLKLKDWPPSDKFEDLLPRHCEEFIRFLPFQQYTDPRAGTLNLAVKLPAHVL 360
Query: 1213 KPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKK 1272
KPD+GPKTYIAYG EEL RGDSVTKLHCD+SDAVNILTHT EV Q I L++
Sbjct: 361 KPDMGPKTYIAYGIREELGRGDSVTKLHCDMSDAVNILTHTAEVLLTDRQKSTISNLKEA 540
Query: 1273 YEAEDMRD 1280
+ A+D R+
Sbjct: 541 HRAQDERE 564
>TC81630 similar to PIR|T06461|T06461 DNA-binding protein PD3 chloroplast -
garden pea, partial (12%)
Length = 1061
Score = 218 bits (554), Expect = 2e-56
Identities = 133/321 (41%), Positives = 164/321 (50%), Gaps = 7/321 (2%)
Frame = +2
Query: 1228 EELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDLYGRINK 1287
EEL RGDSVTKLH D+SDAVN+LTHT +V WQ + I KL+K Y+ ED DLY
Sbjct: 8 EELGRGDSVTKLHLDVSDAVNVLTHTNKVNIAPWQRESINKLKKGYDKEDYSDLY----- 172
Query: 1288 TVVSHRSKHKKCRTGISMDPKIPENDDTMGRNSNLRGSQSNEEIVVNELSTRSSSLGESR 1347
C ++D K S E VN + TRSS + +
Sbjct: 173 -----------CEASANVDGK---------SKSKALDHDQKAENEVNRI-TRSSQVDQ-- 283
Query: 1348 SDSAACVQGFSES----SESKSVLNAGEQAILNMYKRFVKFDLNNHDSGYLFPGKDCEWM 1403
C+ SE ES++ + +
Sbjct: 284 -----CISSISEDWCGKLESRNTIQCDD-------------------------------- 352
Query: 1404 HYDVNNGKQWCSSPV---MPCSKIQIDKTVPVKNDISSNNFFQNDDHMETQFGSAVWDIF 1460
NGK C+ + + D + K + ++ D+ E G AVWDIF
Sbjct: 353 -----NGKGSCTYRMRINFSDGNVSSDPKIESKQGMGRDSL-DIDNGAEAVLGGAVWDIF 514
Query: 1461 RRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQILYLNEKHKRQLKKEYGIEPWTF 1520
RRQDVPKL EYL+KH KEFRH NN PVDSV HPIHDQ L+LNE+HK+QLK+E+ +EPWTF
Sbjct: 515 RRQDVPKLIEYLRKHKKEFRHINNEPVDSVIHPIHDQTLFLNERHKKQLKREFNVEPWTF 694
Query: 1521 EQHLGEAVFIPAGCPHQVRNR 1541
EQHLGEAVFIPAGCPHQVRNR
Sbjct: 695 EQHLGEAVFIPAGCPHQVRNR 757
>BE204827 similar to PIR|F86222|F86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (18%)
Length = 626
Score = 159 bits (401), Expect = 1e-38
Identities = 83/209 (39%), Positives = 117/209 (55%), Gaps = 11/209 (5%)
Frame = +3
Query: 816 KCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKDISVMTGSGEADTGVIL 875
KC R+ YC CI+ WY +EI+ CP C CNC++CL+ D S+ E L
Sbjct: 6 KCDRRGYCDSCISTWYSDIPLDEIQKICPACRGICNCKICLRSDNSIKVRIREIPVLDKL 185
Query: 876 QKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPL--------MEEDIQFDNCNTSIVN 927
Q L LL+ LP+++ I REQ E+E+E + G+ + +E + + C I +
Sbjct: 186 QYLHVLLSSVLPVVKQIHREQCFEVELEKKLRGAEIDLPRTKLNADEQMCCNLCRIPITD 365
Query: 928 FHRSCPNPNCRYDLCLTCCMELRNG-LHYEDIPASGNEETIDEPPITS--AWRAEINGRI 984
+HR C P+C YDLCL CC +LR LH + P + + +T D ++ WR+ NG I
Sbjct: 366 YHRRC--PSCSYDLCLICCRDLREATLHQSEEPQTEHAKTTDRNILSKFPHWRSNDNGSI 539
Query: 985 PCPPKARGGCGTSILSLRRLFEANWVNKL 1013
PCPPK GGCG S L+L R+F+ NWV KL
Sbjct: 540 PCPPKEYGGCGYSSLNLSRIFKMNWVAKL 626
>BI271152 weakly similar to PIR|T05151|T051 hypothetical protein F18E5.50 -
Arabidopsis thaliana, partial (10%)
Length = 511
Score = 134 bits (337), Expect = 3e-31
Identities = 73/145 (50%), Positives = 91/145 (62%), Gaps = 1/145 (0%)
Frame = +1
Query: 1132 CLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPF 1191
CLDWCEV+INI Q+F G L+ R RN W EMLKL W S F+E FP H +E I LP
Sbjct: 79 CLDWCEVEINIRQYFTGSLKCRPQRNTWHEMLKLNGWLSSQVFKEQFPAHFSEVIDALPV 258
Query: 1192 SDYTHPKFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNIL 1250
+Y +P G+LNLA LP K D+GP YI+YG + + DSVTKL CD D VNI+
Sbjct: 259 QEYMNPVSGLLNLAANLPDRSPKHDIGPYVYISYGCAD--TEADSVTKLCCDSYDVVNIM 432
Query: 1251 THTEEVKAPLWQPKIIKKLQKKYEA 1275
TH+ +V Q I+KL KK++A
Sbjct: 433 THSADVPLSTEQLTKIRKLLKKHKA 507
>AW981209 similar to PIR|F86222|F86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (12%)
Length = 753
Score = 120 bits (300), Expect = 5e-27
Identities = 53/98 (54%), Positives = 73/98 (74%)
Frame = +3
Query: 1443 QNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQILYLN 1502
+N D E +WD+FRRQDVPK+TEYLK H KEF +++ D VT P++ ++L+
Sbjct: 354 ENGDVSEITHPGVLWDVFRRQDVPKVTEYLKMHWKEFGNSD----DIVTWPLYGGAIFLD 521
Query: 1503 EKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
HKR+LK+E+G+EPW+FEQ+LGEA+F+PAGCP Q RN
Sbjct: 522 RHHKRKLKEEFGVEPWSFEQNLGEAIFVPAGCPFQARN 635
Score = 58.2 bits (139), Expect = 2e-08
Identities = 40/120 (33%), Positives = 64/120 (53%), Gaps = 6/120 (5%)
Frame = +3
Query: 1222 IAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDL 1281
I+YG +EL RGDSVTKLH ++ D V +L H+ EVK WQ ++ +QK + + ++
Sbjct: 3 ISYGISDELGRGDSVTKLHFNMRDMVYLLVHSSEVKLKDWQRTNVEMMQKTSKESEEKES 182
Query: 1282 YGRINKTVVSHRSK-HKKCRTGIS-MDPKIPENDDTMGRN----SNLRGSQSNEEIVVNE 1335
+G + + S S T I+ +D + + D TM + S+ G+ N EI + E
Sbjct: 183 HG--DPDICSRASSPDSSFYTKINGLDLESDQKDSTMDQGVEVYSSAEGNLVNSEIPLRE 356
>CB895025 similar to PIR|F86222|F862 hypothetical protein [imported] -
Arabidopsis thaliana, partial (1%)
Length = 788
Score = 102 bits (253), Expect = 1e-21
Identities = 71/247 (28%), Positives = 104/247 (41%), Gaps = 43/247 (17%)
Frame = +2
Query: 835 TREEIEIACPFCLRNCNCRLCL------KKDISVMTGSGEADTGVILQKLLYLLNKTLPL 888
T+ E++ ACP C C+C+ C ++ + G+ D + YL+ LP+
Sbjct: 8 TQNEVKKACPVCRGTCSCKDCRASQCKDRESKDCLAGTSRVDR---ILHFHYLVCMLLPV 178
Query: 889 LQHIQREQISEMEVEASMHG---SPLMEEDIQFD--------NCNTSIVNFHRSCPNPNC 937
++ I +Q +E+E EA G S ++ + I+FD C T I+N HRSC N C
Sbjct: 179 IKQISEDQHAELETEAKNKGESISDIIIKQIEFDCNEIIDCNYCKTPILNLHRSCLN--C 352
Query: 938 RYDLCLTCCMELRNGLHYEDIPA-------------------------SGNEETIDEPPI 972
Y LCL CC L G +E I + S ++ET +
Sbjct: 353 SYSLCLRCCQTLSQGSPFEHINSPLTELPDKMDTCIADESCLFEDKSISSDDETDTSMLL 532
Query: 973 TSAWRAEINGRIPCPPKARGGCGTSILSLRRLFEANWVNKLVRNAEELTIQYHPPSV-DL 1031
S I CPP GGCG L LR +F +W+ + AEE+ Y P + D
Sbjct: 533 DSTGFNGTTDSISCPPSELGGCGNDNLDLRCVFPISWIEDMEAKAEEIVCSYDVPEILDK 712
Query: 1032 LVGCLQC 1038
C C
Sbjct: 713 NSSCSLC 733
>TC82777 weakly similar to PIR|D85438|D85438 hypothetical protein AT4g37110
[imported] - Arabidopsis thaliana, partial (11%)
Length = 673
Score = 43.5 bits (101), Expect = 6e-04
Identities = 25/74 (33%), Positives = 37/74 (49%), Gaps = 6/74 (8%)
Frame = +3
Query: 799 RCHQCWRNSWSGVVICAKCKRKQ--YCYECITKWYPGKTREEIEI----ACPFCLRNCNC 852
+CHQC R + + + C KC+ Q C +C+ Y G+ E I CP C CNC
Sbjct: 27 KCHQCGRLTVAQLTDCNKCELPQGRLCGDCLYTRY-GENVTEANINPKWTCPSCREICNC 203
Query: 853 RLCLKKDISVMTGS 866
C +K+ + TG+
Sbjct: 204 NSCRRKNGWLPTGN 245
>AI974510
Length = 334
Score = 42.4 bits (98), Expect = 0.001
Identities = 24/69 (34%), Positives = 33/69 (47%), Gaps = 3/69 (4%)
Frame = +3
Query: 973 TSAWRAEING--RIPCPPKARGGCGTSILSLRRLFEANWVNKLVRNAEELTIQY-HPPSV 1029
TS R N ++ CPP GGCGT +L L +F + + K+ AEE+ Y P +
Sbjct: 123 TSPERTNCNDIEKVSCPPTELGGCGTGLLDLLCIFPSTLLRKMEVKAEEIVCSYDFPETS 302
Query: 1030 DLLVGCLQC 1038
D C C
Sbjct: 303 DKSSSCSLC 329
>AW688542 similar to PIR|T05151|T051 hypothetical protein F18E5.50 -
Arabidopsis thaliana, partial (6%)
Length = 542
Score = 36.6 bits (83), Expect = 0.077
Identities = 19/64 (29%), Positives = 29/64 (44%)
Frame = +1
Query: 10 RCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRSGRRKPVDNSE 69
RCGR WRC R + +CE H+L Q K +++E +R + K + +
Sbjct: 127 RCGRTDGKQWRCKRRVMDNLKLCEIHYL--QGKHRQYREKVPESLKLQRKRKNKEEEQEQ 300
Query: 70 NGVV 73
VV
Sbjct: 301 ETVV 312
>TC80273 similar to GP|8978267|dbj|BAA98158.1 contains similarity to AT-hook
DNA-binding protein~gene_id:K2I5.6 {Arabidopsis
thaliana}, partial (53%)
Length = 1418
Score = 28.9 bits (63), Expect(2) = 0.18
Identities = 14/25 (56%), Positives = 15/25 (60%)
Frame = +3
Query: 452 NNEGSGAGEIVRPKKRGRPRGSKNK 476
N G GA V + RGRP GSKNK
Sbjct: 375 NTSGDGATIEVSRRPRGRPPGSKNK 449
Score = 25.0 bits (53), Expect(2) = 0.18
Identities = 18/63 (28%), Positives = 32/63 (50%), Gaps = 4/63 (6%)
Frame = +3
Query: 502 GRPKGSKNKQKNIVQVSQE---VAGSADCEIAGPKKCGRPKGSMKKRKSL-VCASILEGA 557
GRP GSKNK K + ++++ V +I+G +RK++ +C +L G+
Sbjct: 423 GRPPGSKNKPKPPIIITRDPETVMSPFILDISGGNDVVEAISEFSRRKNIGLC--VLTGS 596
Query: 558 GGI 560
G +
Sbjct: 597 GTV 605
>BG455563 similar to PIR|T05151|T051 hypothetical protein F18E5.50 -
Arabidopsis thaliana, partial (7%)
Length = 663
Score = 34.7 bits (78), Expect = 0.29
Identities = 18/50 (36%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Frame = +3
Query: 2 DETGEECR----RCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWK 47
+ T EEC RC R WRC RA+ +CE H L Q + ++ K
Sbjct: 102 NSTKEECPPDNLRCSRTDGRQWRCKRRAMENVKLCEVHHLQLQHRQKKVK 251
>TC79938 similar to GP|17933299|gb|AAL48232.1 AT4g17800/dl4935c {Arabidopsis
thaliana}, partial (63%)
Length = 1259
Score = 34.3 bits (77), Expect = 0.38
Identities = 30/114 (26%), Positives = 51/114 (44%), Gaps = 4/114 (3%)
Frame = +2
Query: 452 NNEGSGAGEIVRPKKRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQ 511
N S E R G S N+ ++ + + ++ G + G + GRP GSKNK
Sbjct: 80 NQHDSEEQESNRASVGGGAPFSSNEEDDRSQGLELGSAAGPGDVVGRRPRGRPPGSKNKA 259
Query: 512 KNIVQVSQEVAGSADC---EIAGPKKCGRPKGSMKKRKSL-VCASILEGAGGIT 561
K V +++E A + E+AG + +R+ +C +L G+G +T
Sbjct: 260 KPPVIITRESANTLRAHILEVAGGSDVFECVSTYARRRQRGIC--VLSGSGTVT 415
>TC88820 homologue to PIR|D84890|D84890 probable AT-hook DNA-binding protein
[imported] - Arabidopsis thaliana, partial (58%)
Length = 1544
Score = 33.1 bits (74), Expect = 0.85
Identities = 33/118 (27%), Positives = 51/118 (42%), Gaps = 12/118 (10%)
Frame = +3
Query: 456 SGAGEIVRPKKRGR--------PRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGS 507
SG G + R +KR R P G + K + +A GG G + GRP GS
Sbjct: 612 SGNGSLSRGQKRERNNEDGNNTPTGGEGKDDG----GSGSAGGGSGGEMGRRPRGRPAGS 779
Query: 508 KNKQKNIVQVSQEVAGSADCEIA----GPKKCGRPKGSMKKRKSLVCASILEGAGGIT 561
KNK K + ++++ A + + G ++R+ VC IL G+G +T
Sbjct: 780 KNKPKPPIIITRDSANALRSHVMEVANGCDIMESVTVFARRRQRGVC--ILSGSGTVT 947
>TC85062 similar to EGAD|119543|127780 hypothetical protein F49C5.k
{Caenorhabditis elegans}, partial (5%)
Length = 781
Score = 33.1 bits (74), Expect = 0.85
Identities = 55/235 (23%), Positives = 86/235 (36%), Gaps = 2/235 (0%)
Frame = -3
Query: 199 EEPGFGSFQALLCKDRGCAE-DVIFIGDVTGFEGLSGENTHGFRDEVGGFVENPCFEGEN 257
E+ S ALL D E D + GD + + + +G R + G N E
Sbjct: 584 EKQAIVSETALLKNDLASVEGDDVAAGDSSSIQVI---RKYGRRKKKPGRKSNAEIE--- 423
Query: 258 DSNKEGPGSNYKMSALGFEEEIGLLLSRGGTTNEEARCEALRPLSKRGRPKGSKNENDNK 317
KE G+ K ++++ + G + + A +A P KRGR + ++ EN+
Sbjct: 422 ---KEKIGNGTKEETSKGDDDV----ADGDSISVHASTDATTPAKKRGRKRNTEKENE-- 270
Query: 318 QLSTALDGQSVGGDDNAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAE-KSG 376
T D + GG + G + K D V+DL E R + K G
Sbjct: 269 ---TGKDAEVEGGFSSVGVRKSERPRKI----------KSLKEDYVSDLEEDVRKKGKRG 129
Query: 377 RPKVSKNKIRRVEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEAGDEAAGEIGG 431
R KV ++ V K + K +K + +GDE E G
Sbjct: 128 RKKV-------------IIGVSDENVKTEKKQPGRK--RKELFSSGDENEAENEG 9
>BQ137244 similar to GP|20161222|dbj Epstein-Barr virus EBNA-1-like protein
{Oryza sativa (japonica cultivar-group)}, partial (5%)
Length = 1050
Score = 33.1 bits (74), Expect = 0.85
Identities = 20/68 (29%), Positives = 33/68 (48%)
Frame = +1
Query: 675 KRGRPKGSKNKMKSIANKARNKFGKVRNMRGRPKGSLRKKNETAYCLDSQNERNSLDGRT 734
+RG+P+ S + ARN G+ R+ G + R++NE+ D+ R T
Sbjct: 697 ERGQPRASARTR----DDARNSSGRDRDASGEERRERRERNESGRARDAIESRPDATNGT 864
Query: 735 STEAAYRN 742
+ EAA R+
Sbjct: 865 AREAARRD 888
>TC78237 similar to GP|17933299|gb|AAL48232.1 AT4g17800/dl4935c {Arabidopsis
thaliana}, partial (59%)
Length = 1166
Score = 33.1 bits (74), Expect = 0.85
Identities = 16/50 (32%), Positives = 29/50 (58%)
Frame = +3
Query: 440 GSLNKLKNTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNNIMEVSKKAAS 489
G+ N +D + G G++V + RGRP GSKNK + +++++A+
Sbjct: 192 GNNNNNHEGLDLVSPNHGLGDVVGRRPRGRPPGSKNKPKPPVIITRESAN 341
Score = 32.0 bits (71), Expect = 1.9
Identities = 26/91 (28%), Positives = 42/91 (45%), Gaps = 4/91 (4%)
Frame = +3
Query: 475 NKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSADCEI----A 530
N NN + + + G + G + GRP GSKNK K V +++E A + I +
Sbjct: 195 NNNNNHEGLDLVSPNHGLGDVVGRRPRGRPPGSKNKPKPPVIITRESANTLRAHILEVSS 374
Query: 531 GPKKCGRPKGSMKKRKSLVCASILEGAGGIT 561
G +KR+ +C +L G+G +T
Sbjct: 375 GCDVFDSVATYARKRQRGIC--VLSGSGTVT 461
>TC91283 weakly similar to GP|21554159|gb|AAM63238.1 unknown {Arabidopsis
thaliana}, partial (31%)
Length = 784
Score = 32.7 bits (73), Expect = 1.1
Identities = 28/92 (30%), Positives = 40/92 (43%), Gaps = 3/92 (3%)
Frame = +2
Query: 473 SKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQE---VAGSADCEI 529
S VN+ +E S SG +G + GRP GSKNK K + +++E S E+
Sbjct: 401 SNGHVNDELENSN-GRSGDQTARSGRRPRGRPPGSKNKPKPPLMITKETPNALSSVILEV 577
Query: 530 AGPKKCGRPKGSMKKRKSLVCASILEGAGGIT 561
A S R+ S+L G G +T
Sbjct: 578 ANGADIAHSISSYANRRHR-GVSVLSGTGYVT 670
>TC77972 similar to GP|6175162|gb|AAF04888.1| hypothetical protein
{Arabidopsis thaliana}, partial (46%)
Length = 1589
Score = 32.7 bits (73), Expect = 1.1
Identities = 21/75 (28%), Positives = 33/75 (44%), Gaps = 11/75 (14%)
Frame = +2
Query: 624 GEEQKDHGSDIGKP-IGLDNDKATLASDRDQETPNQTLAQDEVQND----------KSSV 672
G + S +GKP +G +++ + + N +DE +N +S
Sbjct: 374 GMDNSVTSSPLGKPDLGFSMNQSAVTGVNNMNNNNNEEEEDEKENSDEHKGGAIETNTST 553
Query: 673 KPKRGRPKGSKNKMK 687
+ RGRP GSKNK K
Sbjct: 554 RRPRGRPSGSKNKPK 598
>TC81253 similar to GP|9757941|dbj|BAB08429.1 gene_id:MJC20.6~unknown
protein {Arabidopsis thaliana}, partial (50%)
Length = 1032
Score = 31.6 bits (70), Expect = 2.5
Identities = 23/69 (33%), Positives = 36/69 (51%), Gaps = 7/69 (10%)
Frame = +1
Query: 393 NVVAVKIVGPKKHGR-------PKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNKL 445
NV+ + KK GR K SK KKN++ A +E + +IGG + RP +L
Sbjct: 610 NVMDQQSSSSKKKGRLNRHQEMSKGSKNEKKNLLSA-EEPSRDIGGWEDSQRPVEYQQRL 786
Query: 446 KNTVDCNNE 454
+T+D ++E
Sbjct: 787 HDTIDGDSE 813
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,495,449
Number of Sequences: 36976
Number of extensions: 753575
Number of successful extensions: 3128
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 2934
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3036
length of query: 1541
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1432
effective length of database: 4,984,343
effective search space: 7137579176
effective search space used: 7137579176
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0103.12


