

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100c.1
(773 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
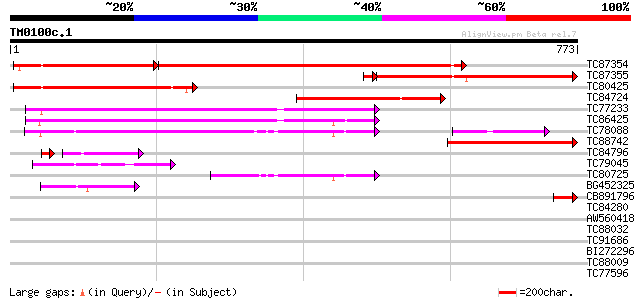
Score E
Sequences producing significant alignments: (bits) Value
TC87354 similar to GP|2576411|gb|AAC61784.1| similar to dynamin-... 682 0.0
TC87355 similar to GP|2576411|gb|AAC61784.1| similar to dynamin-... 394 e-113
TC80425 similar to GP|19386736|dbj|BAB86118. putative dynamin-li... 374 e-104
TC84724 similar to PIR|T04982|T04982 dynamin-like protein ADL2 -... 335 4e-92
TC77233 homologue to PIR|S63668|S63668 phragmoplastin 5 - soybea... 323 2e-88
TC86425 similar to GP|18854996|gb|AAL79688.1 putative phragmopla... 310 1e-84
TC78088 similar to GP|11991506|emb|CAC19656. dynamin-like protei... 304 1e-82
TC88742 similar to PIR|T04982|T04982 dynamin-like protein ADL2 -... 278 6e-75
TC84796 similar to PIR|E96630|E96630 hypothetical protein F8A5.7... 74 2e-15
TC79045 homologue to GP|6625788|gb|AAF19398.1| dynamin homolog {... 74 3e-13
TC80725 similar to GP|18854996|gb|AAL79688.1 putative phragmopla... 63 4e-10
BG452325 similar to GP|9294439|dbj gene_id:MMB12.21~unknown prot... 53 4e-07
CB891796 48 2e-05
TC84280 weakly similar to PIR|T48778|T48778 hypothetical protein... 34 0.19
AW560418 similar to GP|6625788|gb| dynamin homolog {Astragalus s... 33 0.32
TC88032 similar to GP|9294381|dbj|BAB02391.1 gene_id:MIE1.9~unkn... 32 1.2
TC91686 similar to GP|22136246|gb|AAM91201.1 unknown protein {Ar... 31 1.6
BI272296 similar to GP|12322014|gb GTPase putative; 34281-30152... 31 2.1
TC88009 similar to PIR|T48507|T48507 probable GTP-binding protei... 30 4.6
TC77596 similar to GP|18377702|gb|AAL67001.1 putative clathrin b... 30 4.6
>TC87354 similar to GP|2576411|gb|AAC61784.1| similar to dynamin-like protein
encoded by GenBank Accession Number X99669 {Arabidopsis
thaliana}, partial (65%)
Length = 1971
Score = 682 bits (1759), Expect(2) = 0.0
Identities = 348/422 (82%), Positives = 378/422 (89%), Gaps = 2/422 (0%)
Frame = +1
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQM 262
DALQMA VADP+GNRTIGVITKLDIMDRGTDARNLL GKVIPLRLGYVGVVNRSQEDIQM
Sbjct: 706 DALQMAGVADPEGNRTIGVITKLDIMDRGTDARNLLQGKVIPLRLGYVGVVNRSQEDIQM 885
Query: 263 NRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSL 322
NRSIKDALVAEE+FFRSRPVYSGLADSCG+PQLAKKLN+ILAQHIKA+LPGL+A IST+L
Sbjct: 886 NRSIKDALVAEEKFFRSRPVYSGLADSCGIPQLAKKLNQILAQHIKALLPGLRAHISTNL 1065
Query: 323 VALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYI 382
V +AKEHASYGEITESKAGQGAL+LNILSKYC+AFSSM++GKN+EMST ELSGGARIHYI
Sbjct: 1066 VTVAKEHASYGEITESKAGQGALLLNILSKYCEAFSSMVDGKNEEMSTSELSGGARIHYI 1245
Query: 383 FQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSL 442
FQSI+VRSLE VDPCE LTDDDIRTA+QNATGP+SALF+PEVPFE+ VRRQISRLLDPSL
Sbjct: 1246 FQSIFVRSLEEVDPCEGLTDDDIRTAIQNATGPRSALFVPEVPFEVLVRRQISRLLDPSL 1425
Query: 443 QCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMD 502
QCARFIYDEL+K+SHRCM TELQRFPFLRK MDEVIGNFLR+GLEPSE MI HI+EMEMD
Sbjct: 1426 QCARFIYDELIKMSHRCMVTELQRFPFLRKRMDEVIGNFLREGLEPSENMIAHIMEMEMD 1605
Query: 503 YINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LDALESDKGLASERNGKSRGIIARQ 560
YINTSHPNFIGGSKALE AVQQTK SR + +S DALES+KG ASER KSR I+ARQ
Sbjct: 1606 YINTSHPNFIGGSKALEIAVQQTKSSRAALSLSRPKDALESEKGSASERTVKSRAILARQ 1785
Query: 561 ANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNRVYVKENTSTKPHAEPVHN 620
ANGVV D VR SD EK+A SGN GGSSW ISSIFGG D EN +K HAEP H+
Sbjct: 1786 ANGVVVDPAVRAVSDAEKIASSGNIGGSSWGISSIFGGGDKS--AXENVXSKQHAEPFHS 1959
Query: 621 VQ 622
V+
Sbjct: 1960 VE 1965
Score = 308 bits (788), Expect(2) = 0.0
Identities = 161/201 (80%), Positives = 179/201 (88%), Gaps = 4/201 (1%)
Frame = +3
Query: 6 DEAVTAA----STAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSS 61
D+ VT+ S+A PLGS VISLVNRLQDIFSRVG+QS I +LPQVAVVGSQSSGKSS
Sbjct: 105 DDTVTSPPPPPSSAPVPLGSGVISLVNRLQDIFSRVGSQSAI-NLPQVAVVGSQSSGKSS 281
Query: 62 VLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQA 121
VLE+LVGRDFLPRG +ICTRRPLVLQLV + E+GEFLHLPG++F+DF+ IR EIQA
Sbjct: 282 VLEALVGRDFLPRGNEICTRRPLVLQLVRSSEPADEYGEFLHLPGKRFYDFSDIRREIQA 461
Query: 122 ETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSY 181
ETDREAG+NKGVS++QIRLKI SPNVLD+TLVDLPGITKVPVGDQPSDIEARIRTMIMSY
Sbjct: 462 ETDREAGDNKGVSDRQIRLKIVSPNVLDMTLVDLPGITKVPVGDQPSDIEARIRTMIMSY 641
Query: 182 IKIPTCLILAVTPANSDLANS 202
IK P+CLILAVTPANSDLANS
Sbjct: 642 IKEPSCLILAVTPANSDLANS 704
>TC87355 similar to GP|2576411|gb|AAC61784.1| similar to dynamin-like
protein encoded by GenBank Accession Number X99669
{Arabidopsis thaliana}, partial (23%)
Length = 1317
Score = 394 bits (1012), Expect(2) = e-113
Identities = 206/279 (73%), Positives = 231/279 (81%), Gaps = 6/279 (2%)
Frame = +1
Query: 501 MDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSL--DALESDKGLASERNGKSRGIIA 558
MDYINTSHPNFIGGSKALE AVQQTK SR + +S DALES+KG ASER KSR I+A
Sbjct: 148 MDYINTSHPNFIGGSKALEIAVQQTKSSRAALSLSRPKDALESEKGSASERTVKSRAILA 327
Query: 559 RQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNRVYVKENTSTKPHAEPV 618
RQANGVV D VR SD EK+A SGN GGSSW ISSIFGG D +EN ++K HAEP
Sbjct: 328 RQANGVVVDPAVRAVSDAEKIASSGNIGGSSWGISSIFGGGDKSA--RENVASKQHAEPF 501
Query: 619 HNV---QSSSTIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYYDIARKNIEDLVPKA 675
H+V QS S IHL EPP +LRPS+SNSE E VEITVTKLLL+SYYDI RKN+ED VPKA
Sbjct: 502 HSVEVEQSFSMIHLTEPPTILRPSDSNSETEAVEITVTKLLLKSYYDIVRKNVEDFVPKA 681
Query: 676 IMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRELLRAYQQAFRDLDE 735
IMHFLVNNTKRELHNVFI+KLYR++LFEEMLQEP E+A+KRKRCRELLRAYQQAF+DL+E
Sbjct: 682 IMHFLVNNTKRELHNVFIKKLYRDNLFEEMLQEPDEIASKRKRCRELLRAYQQAFKDLEE 861
Query: 736 LPLEAETVERGYGSPEATGLPRIRGLPSSSMYS-SSLGD 773
LP+EAET+ERGY PE TGLP+I GLP+SSMYS +S GD
Sbjct: 862 LPMEAETLERGYSLPETTGLPKIHGLPTSSMYSTNSSGD 978
Score = 33.9 bits (76), Expect(2) = e-113
Identities = 14/19 (73%), Positives = 17/19 (88%)
Frame = +2
Query: 483 RDGLEPSETMITHIIEMEM 501
R+GLEPSE MI HI+EME+
Sbjct: 2 REGLEPSENMIAHIMEMEI 58
>TC80425 similar to GP|19386736|dbj|BAB86118. putative dynamin-like protein
ADL2 {Oryza sativa (japonica cultivar-group)}, partial
(29%)
Length = 885
Score = 374 bits (960), Expect = e-104
Identities = 201/253 (79%), Positives = 216/253 (84%), Gaps = 3/253 (1%)
Frame = +3
Query: 6 DEAVTAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLES 65
++ + A S PLGS+VISLVN+LQDIFSRVG+QSTI DLPQVAVVG QSSGKSSVLE+
Sbjct: 135 NQTMAAVSNDGAPLGSTVISLVNKLQDIFSRVGSQSTI-DLPQVAVVGCQSSGKSSVLEA 311
Query: 66 LVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDR 125
LVGRDFLPRG DICTRRPLVLQLVH PS PE EFLHLPGR FHDF+QIR EIQAETDR
Sbjct: 312 LVGRDFLPRGNDICTRRPLVLQLVHIPPSKPESAEFLHLPGRTFHDFSQIRAEIQAETDR 491
Query: 126 EAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIP 185
EAG NKGVS+KQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIE+RIRTMIMSYIK+P
Sbjct: 492 EAGGNKGVSDKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIESRIRTMIMSYIKVP 671
Query: 186 TCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLL---GKV 242
TCLILAVTPANSDLANSDALQMA ADPDG+RTI + GT+ L + GKV
Sbjct: 672 TCLILAVTPANSDLANSDALQMAGNADPDGHRTIR--CHYQVGYHGTEVPMLGICYWGKV 845
Query: 243 IPLRLGYVGVVNR 255
IPLRLGYVGVVNR
Sbjct: 846 IPLRLGYVGVVNR 884
>TC84724 similar to PIR|T04982|T04982 dynamin-like protein ADL2 -
Arabidopsis thaliana, partial (17%)
Length = 608
Score = 335 bits (859), Expect = 4e-92
Identities = 168/204 (82%), Positives = 182/204 (88%)
Frame = +3
Query: 391 LEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYD 450
LE VDPCEDLTDDDIRTA+QNATGP++ALF+P+VPFE+ VRRQISRLLDPSLQCARFIYD
Sbjct: 3 LEEVDPCEDLTDDDIRTAIQNATGPRAALFVPDVPFEVLVRRQISRLLDPSLQCARFIYD 182
Query: 451 ELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSHPN 510
ELMKISHRCM TELQRFPFLRK MDEVIGNFLR+GLEPSETMITHIIEMEMDYINTSHPN
Sbjct: 183 ELMKISHRCMVTELQRFPFLRKRMDEVIGNFLREGLEPSETMITHIIEMEMDYINTSHPN 362
Query: 511 FIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASERNGKSRGIIARQANGVVADHGV 570
FIGGSKALEAAVQQTK S VS DA+ESDKG ASER+GKSR I+AR ANG +ADHGV
Sbjct: 363 FIGGSKALEAAVQQTKSSTVSK--VKDAVESDKGSASERSGKSRSILARHANGGMADHGV 536
Query: 571 RGASDVEKVAPSGNAGGSSWRISS 594
R +SD++KV SG GSSW ISS
Sbjct: 537 RASSDIDKVLHSGTTSGSSWGISS 608
>TC77233 homologue to PIR|S63668|S63668 phragmoplastin 5 - soybean, complete
Length = 2321
Score = 323 bits (828), Expect = 2e-88
Identities = 191/494 (38%), Positives = 291/494 (58%), Gaps = 11/494 (2%)
Frame = +3
Query: 22 SVISLVNRLQDIFSRVGNQS------TIID-LPQVAVVGSQSSGKSSVLESLVGRDFLPR 74
++ISLVN++Q + +G+ T+ D LP +AVVG QSSGKSSVLES+VG+DFLPR
Sbjct: 288 NLISLVNKIQRACTALGDHGESSALPTLWDSLPAIAVVGGQSSGKSSVLESVVGKDFLPR 467
Query: 75 GTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVS 134
G+ I TRRPLVLQL + E+ EFLHLP ++F DF +R EIQ ETDRE G +K +S
Sbjct: 468 GSGIVTRRPLVLQLHKSDEGTREYAEFLHLPRKRFTDFAAVRKEIQDETDRETGRSKQIS 647
Query: 135 EKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTP 194
I L I+SPNV+++TL+DLPG+TKV V QP I I M+ SYI+ P C+ILA++P
Sbjct: 648 TVPIHLSIYSPNVVNLTLIDLPGLTKVAVDGQPESIVHDIENMVRSYIEKPNCIILAISP 827
Query: 195 ANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVN 254
AN D+A SDA+++++ DP G RTIGV+TK+D+MD+GTDA ++L G+ L+ ++GVVN
Sbjct: 828 ANQDIATSDAIKISREVDPTGERTIGVLTKIDLMDKGTDAVDMLEGRAYRLKFPWIGVVN 1007
Query: 255 RSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGL 314
RSQ DI N + A E E+F S P Y LA+ G LAK L+K L IK+ +PG+
Sbjct: 1008RSQADINKNVDMIAARRREREYFNSTPEYKHLANRMGSEHLAKMLSKHLETVIKSKIPGI 1187
Query: 315 KARISTSLVALAKEHASYGEITESKAGQGAL--VLNILSKYCDAFSSMLEGKNKEMSTCE 372
++ IS ++ + E + G+ + AG G L ++ I + F L+G
Sbjct: 1188QSLISKTINEIETELSRLGKPVAADAG-GKLYAIMEICRSFDQIFKDHLDGVR------- 1343
Query: 373 LSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRR 432
GG +I+ +F + +L+ + + L+ ++IR + A G + L PE + +
Sbjct: 1344-PGGDKIYNVFDNQLPAALKRLQFDKQLSMENIRKLITEADGYQPHLIAPEQGYRRLIES 1520
Query: 433 QISRLLDPSLQCARFIYDELMKISHRCM--FTELQRFPFLRKHMDEVIGNFLRDGLEPSE 490
+ + P+ ++ L + H+ + +L+++P LR + + L + S+
Sbjct: 1521TLVTIRGPAEAAVDAVHSLLKDLVHKAVSETLDLKQYPGLRAEVGAAAIDSLDRMRDESK 1700
Query: 491 TMITHIIEMEMDYI 504
+++ME Y+
Sbjct: 1701RATLQLVDMESGYL 1742
>TC86425 similar to GP|18854996|gb|AAL79688.1 putative phragmoplastin {Oryza
sativa}, partial (98%)
Length = 2328
Score = 310 bits (795), Expect = 1e-84
Identities = 194/497 (39%), Positives = 276/497 (55%), Gaps = 14/497 (2%)
Frame = +1
Query: 22 SVISLVNRLQDIFSRVGN----------QSTIIDLPQVAVVGSQSSGKSSVLESLVGRDF 71
S+I LVNR+Q +++G+ S LP VAVVG QSSGKSSVLES+VGRDF
Sbjct: 181 SLIGLVNRIQQACTKLGDYGGSDSNNTFSSLWEALPSVAVVGGQSSGKSSVLESIVGRDF 360
Query: 72 LPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENK 131
LPRG+ I TRRPLVLQL + E+ EFLH PGRK DF +R EIQ ETDR G+ K
Sbjct: 361 LPRGSGIVTRRPLVLQLHKIEDGEKEYAEFLHRPGRKITDFAMVRQEIQDETDRITGKTK 540
Query: 132 GVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILA 191
+S I L I+SPNV+++TL+DLPG+TKV V Q I I M+ S+I P C+ILA
Sbjct: 541 QISPIPIHLSIYSPNVVNLTLIDLPGLTKVAVEGQSESIVEDIENMVRSFIDKPNCIILA 720
Query: 192 VTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVG 251
++PAN D+A SDA+++++ DP G RT GV+TKLD+MD+GT+A ++L G+ L+ +VG
Sbjct: 721 ISPANQDIATSDAIKISREVDPSGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPWVG 900
Query: 252 VVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVL 311
VVNRSQ DI N + A E E+F + P Y LA G LAK L++ L I+A +
Sbjct: 901 VVNRSQADINKNTDMIVARRKEVEYFETSPDYGHLASKMGSEYLAKLLSQHLESVIRARI 1080
Query: 312 PGLKARISTSLVALAKEHASYGEITESKAG-QGALVLNILSKYCDAFSSMLEGKNKEMST 370
P + + I+ S+ L E G AG Q +L + K+ F L+G
Sbjct: 1081PSITSLINKSIEELESEMDHLGRPIAVDAGAQLYTILELCRKFERVFKEHLDGGR----- 1245
Query: 371 CELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFV 430
GG RI+ +F + +L + + L+ +++ + A G + L PE + +
Sbjct: 1246---PGGDRIYNVFDNQLPAALRKLPIDKHLSLQNVKRVVSEADGYQPHLIAPEQGYRRLI 1416
Query: 431 RRQISRLLDP---SLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLE 487
+S P S+ F+ EL++ S EL+RFP L+ + L +
Sbjct: 1417EGTLSYFRGPAEASVDAVHFVLKELVRKSIG-ETEELRRFPTLQAELAAATTEALERFRD 1593
Query: 488 PSETMITHIIEMEMDYI 504
S+ +++ME Y+
Sbjct: 1594ESKKTTIRLVDMEASYL 1644
>TC78088 similar to GP|11991506|emb|CAC19656. dynamin-like protein DLP1
{Arabidopsis thaliana}, partial (98%)
Length = 2343
Score = 304 bits (778), Expect = 1e-82
Identities = 193/492 (39%), Positives = 280/492 (56%), Gaps = 8/492 (1%)
Frame = +3
Query: 21 SSVISLVNRLQDIFSRVGNQ-----STIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRG 75
+S+I L+N++Q + +G+ S LP VAVVG QSSGKSSVLES+VGRDFLPRG
Sbjct: 249 TSLIGLINKIQRACTVLGDHGGEGLSLWEALPSVAVVGGQSSGKSSVLESVVGRDFLPRG 428
Query: 76 TDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSE 135
+ I TRRPLVLQL H + E+ EFLHLP ++F DF +R EI ETDR G++K +S
Sbjct: 429 SGIVTRRPLVLQL-HKTENGQEYAEFLHLPRKRFTDFAAVRKEIADETDRITGKSKQISN 605
Query: 136 KQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPA 195
I+L I+SPNV+++TL+DLPG+TKV V Q I I M+ SY++ P C+ILA++PA
Sbjct: 606 IPIQLSIYSPNVVNLTLIDLPGLTKVAVEGQQESIVQDIEQMVRSYVEKPNCIILAISPA 785
Query: 196 NSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNR 255
N D+A SDA+++A+ DP G RT GV+TKLD+MD+GT+A ++L G+ L+ +VG+VNR
Sbjct: 786 NQDIATSDAIKIAKEVDPSGERTFGVVTKLDLMDKGTNAVDVLEGRQYRLQHPWVGIVNR 965
Query: 256 SQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLK 315
SQ DI N + A E E+F + P Y LA G LA+ L++ L Q I+ +P +
Sbjct: 966 SQADINKNVDMIVARRKEREYFETSPEYGHLAHKMGSEYLARLLSQHLEQVIRQKIPSII 1145
Query: 316 ARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSG 375
A I+ ++ L E G GA + IL + C AF + KE G
Sbjct: 1146ALINKTIDELNAELDRIGR--PIAVDSGAQLYTIL-EMCRAFDKVF----KEHLDGGRPG 1304
Query: 376 GARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQIS 435
G RI+ +F +L+ + L+ +++ + A G + L PE + + IS
Sbjct: 1305GDRIYGVFDHQLPAALKKLPFDRHLSLKNVQKVVTEADGYQPHLIAPEQGYRRLIEGSIS 1484
Query: 436 RLLDP---SLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETM 492
P S+ F+ EL++ S EL+RFP L + L + S+
Sbjct: 1485YFKGPAEASVDAVHFVLKELVRKS-IAETEELRRFPTLSNDIATAANEALDKFRDESKKT 1661
Query: 493 ITHIIEMEMDYI 504
+T +++ME Y+
Sbjct: 1662VTRLVDMESSYL 1697
Score = 43.1 bits (100), Expect = 4e-04
Identities = 31/135 (22%), Positives = 60/135 (43%), Gaps = 2/135 (1%)
Frame = +3
Query: 604 YVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYYDI 663
Y+ K H EP N + PP ++++ G ++ +Y ++
Sbjct: 1692 YLTAEFFRKIHLEPEKNPNGPPNSNRNAPPNNDNFTDNHLRKIGSNVS-------AYINM 1850
Query: 664 ARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFE--EMLQEPAEVAAKRKRCRE 721
+++ +PKA++H V KR L N F ++ +++ + ML E + KR + +
Sbjct: 1851 VCDTLKNTIPKAVVHCQVREAKRSLLNYFYVQVGKKEKEKLGAMLDEDPSLMEKRNQIAK 2030
Query: 722 LLRAYQQAFRDLDEL 736
L Y+QA D+D +
Sbjct: 2031 RLELYKQARDDIDSV 2075
>TC88742 similar to PIR|T04982|T04982 dynamin-like protein ADL2 -
Arabidopsis thaliana, partial (15%)
Length = 1161
Score = 278 bits (711), Expect = 6e-75
Identities = 141/178 (79%), Positives = 154/178 (86%), Gaps = 1/178 (0%)
Frame = +1
Query: 597 GGRDNRVYVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVTKLL 656
GG DNRV VKENT++K H +PV +V SSTIHLREPP VLRPSE +SE VEI VTKLL
Sbjct: 1 GGGDNRVSVKENTNSKHHNDPVQSVLPSSTIHLREPPTVLRPSERSSETLAVEIAVTKLL 180
Query: 657 LRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKR 716
LRSYY+I RKN+EDL+PKAIMHFLVNNTKRELHNVFI LYR+DLFEEMLQEP E+A KR
Sbjct: 181 LRSYYEIVRKNVEDLIPKAIMHFLVNNTKRELHNVFIANLYRDDLFEEMLQEPNEIAVKR 360
Query: 717 KRCRELLRAYQQAFRDLDELPLEAETVERGYGSPEATGLPRIRGLPSSSMYS-SSLGD 773
KRCRELLRAYQQAFRDLDELPLEAET E G+ SPE TGLP+IRGLP+SSMYS SS GD
Sbjct: 361 KRCRELLRAYQQAFRDLDELPLEAETAEWGHSSPETTGLPKIRGLPTSSMYSTSSSGD 534
>TC84796 similar to PIR|E96630|E96630 hypothetical protein F8A5.7 [imported]
- Arabidopsis thaliana, partial (55%)
Length = 766
Score = 73.9 bits (180), Expect(2) = 2e-15
Identities = 44/111 (39%), Positives = 62/111 (55%), Gaps = 1/111 (0%)
Frame = +2
Query: 73 PRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRK-FHDFTQIRHEIQAETDREAGENK 131
PRG ICTR PLV++L + PE L G++ D + I T+ AG K
Sbjct: 440 PRGQGICTRVPLVMRLQNHLLPQPEL--VLEYNGKQVLTDEANVSDAINTATEELAGTAK 613
Query: 132 GVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYI 182
G+S + L + V D+T+VDLPGIT+VPV QP +I +I+ +IM Y+
Sbjct: 614 GISNTPLTLIVKKNGVPDLTMVDLPGITRVPVHGQPDNIYDQIKDIIMDYL 766
Score = 27.3 bits (59), Expect(2) = 2e-15
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = +3
Query: 44 IDLPQVAVVGSQSSGKS 60
I LP + VVG QSSGKS
Sbjct: 354 IQLPTIVVVGDQSSGKS 404
>TC79045 homologue to GP|6625788|gb|AAF19398.1| dynamin homolog {Astragalus
sinicus}, partial (22%)
Length = 951
Score = 73.6 bits (179), Expect = 3e-13
Identities = 56/199 (28%), Positives = 94/199 (47%), Gaps = 3/199 (1%)
Frame = +3
Query: 31 QDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVH 90
+DI N V +G+ SGKS+VL SL+G LP G + TR P+V+ L
Sbjct: 381 EDIDETSNNSRRPSTFLNVVALGNVGSGKSAVLNSLIGHPVLPTGENGATRAPIVIDL-Q 557
Query: 91 TKPSHPEFGEFLHLPGRKFH-DFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLD 149
S L + + + +RH +Q DR + + + QI+LK+ +
Sbjct: 558 RDTSLSSKSIILQIDNKNQQVSASALRHSLQ---DRLSKASSAKARDQIKLKLRTSTAPP 728
Query: 150 ITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANS--DLANSDALQM 207
+ LVDLPG+ D + + Y + +++ + PA ++A+S AL++
Sbjct: 729 LMLVDLPGL----------DQRIMDESTVSEYAEHNDAILVVIVPAAQAPEIASSRALRL 878
Query: 208 AQVADPDGNRTIGVITKLD 226
A+ D +G R +GVI+K+D
Sbjct: 879 AKEYDGEGTRIVGVISKID 935
>TC80725 similar to GP|18854996|gb|AAL79688.1 putative phragmoplastin {Oryza
sativa}, partial (54%)
Length = 1248
Score = 63.2 bits (152), Expect = 4e-10
Identities = 61/233 (26%), Positives = 99/233 (42%), Gaps = 3/233 (1%)
Frame = +1
Query: 275 EFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGE 334
E+F + P Y LA+ G LAK L+K L I+A +P + + I+ S+ L E G
Sbjct: 1 EYFATSPDYGHLANKMGSEYLAKLLSKHLESVIRARIPSITSLINKSIEELESEMNHLGR 180
Query: 335 ITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAV 394
A GA + IL + C AF + KE GG RI+ +F + +L +
Sbjct: 181 PIALDA--GAQLYTIL-ELCRAFERIF----KEHLDGGRPGGDRIYNVFDNQLPSALRKL 339
Query: 395 DPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDP---SLQCARFIYDE 451
L+ ++R + A G + L PE + + + P S+ F+ E
Sbjct: 340 PFDRHLSLQNVRKVVSEADGYQPHLIAPEHGYRRLIEGSLGYFKGPAEASVDAVNFVLKE 519
Query: 452 LMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYI 504
L++ S EL+RFP + + L E S+ +++ME Y+
Sbjct: 520 LVRKS-IAETQELKRFPTFQAALAAAANEALERFREESKKTTLRLVDMESSYL 675
>BG452325 similar to GP|9294439|dbj gene_id:MMB12.21~unknown protein
{Arabidopsis thaliana}, partial (25%)
Length = 666
Score = 53.1 bits (126), Expect = 4e-07
Identities = 45/142 (31%), Positives = 69/142 (47%), Gaps = 6/142 (4%)
Frame = +3
Query: 42 TIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEF 101
T D P V VVG Q+ GKS+++E+L+G F G TRRP+ L + + H E
Sbjct: 138 TPFDAPAVLVVGHQTDGKSALVEALMGFQFNHVGGGTKTRRPITLHMKY--GPHCESPSC 311
Query: 102 LHL----PGRKFH-DFTQIRHEIQAETDR-EAGENKGVSEKQIRLKIFSPNVLDITLVDL 155
L P H +QI+ I+AE R E S K+I +K+ ++T++D
Sbjct: 312 YLLSDDDPSLSHHMSLSQIQGYIEAENARLERDSCCQFSAKEIIIKVEYKYCPNLTIIDT 491
Query: 156 PGITKVPVGDQPSDIEARIRTM 177
PG+ G + I+A+ R +
Sbjct: 492 PGLVAPAPGRKNRAIQAQARAV 557
>CB891796
Length = 494
Score = 47.8 bits (112), Expect = 2e-05
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 1/33 (3%)
Frame = -3
Query: 742 TVERGYGSPEATGLPRIRGLPSSSMYS-SSLGD 773
T+ERGY PE TGLP+I GLP+SSMYS +S GD
Sbjct: 492 TLERGYSLPETTGLPKIHGLPTSSMYSTNSSGD 394
>TC84280 weakly similar to PIR|T48778|T48778 hypothetical protein 13E11.260
[imported] - Neurospora crassa, partial (12%)
Length = 1058
Score = 34.3 bits (77), Expect = 0.19
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Frame = -3
Query: 372 ELSGGARIHYIFQSIYVRS-LEAVDPCEDLTDDDIRTALQNATGPKSALFIPE 423
E + IHY ++S+ RS L+ PC T DD+ + + GP+ A +P+
Sbjct: 222 ESTFNTNIHYDYESLASRSQLDPTPPCHSETSDDLLSEYRATRGPRMAADLPQ 64
>AW560418 similar to GP|6625788|gb| dynamin homolog {Astragalus sinicus},
partial (18%)
Length = 517
Score = 33.5 bits (75), Expect = 0.32
Identities = 41/169 (24%), Positives = 72/169 (42%), Gaps = 17/169 (10%)
Frame = +2
Query: 198 DLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLG--------- 248
++++S AL++A+ D + RT+GVITK+D T+ ++L + + L G
Sbjct: 20 EISSSRALRVAKEYDSESTRTVGVITKID--QAATEPKSLAAVQALLLNQGPPKTSDIPW 193
Query: 249 ------YVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKI 302
V + + + S++ A AE E +S G PQ KL +I
Sbjct: 194 VALIGQSVSIASAHSHSAAPDTSLETAWRAETETLKS--------ILTGAPQ--TKLGRI 343
Query: 303 -LAQHIKAVLPG-LKARISTSLVALAKEHASYGEITESKAGQGALVLNI 349
L + A + +K R+ T L L + E GQ +++L +
Sbjct: 344 ALVDSLAAQIRNRMKLRLPTLLTGLQGKSQIVQEELVKLGGQWSVLLKV 490
>TC88032 similar to GP|9294381|dbj|BAB02391.1 gene_id:MIE1.9~unknown protein
{Arabidopsis thaliana}, partial (14%)
Length = 1415
Score = 31.6 bits (70), Expect = 1.2
Identities = 14/41 (34%), Positives = 23/41 (55%)
Frame = +2
Query: 605 VKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEM 645
V+ S KP + VHNV S++T+ +EP P S+ ++
Sbjct: 806 VRNVRSPKPDVDVVHNVPSNTTVVNKEPQKAFSPERSSEKI 928
>TC91686 similar to GP|22136246|gb|AAM91201.1 unknown protein {Arabidopsis
thaliana}, partial (26%)
Length = 669
Score = 31.2 bits (69), Expect = 1.6
Identities = 35/147 (23%), Positives = 60/147 (40%), Gaps = 4/147 (2%)
Frame = +1
Query: 13 STAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFL 72
S+ T +S +S+++ I+ G + VA++G QSSGKS+++ L F
Sbjct: 25 SSTVTVFSTSPVSIISSKLQIWLIPGLSYAV-----VAIMGPQSSGKSTLMNHLFHTSFR 189
Query: 73 P----RGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAG 128
RG T+ + + +P I +++ RE G
Sbjct: 190 EMDAFRGRSQTTKGIWIAKCTGIEP-------------------CTIAMDLEGTDGREKG 312
Query: 129 ENKGVSEKQIRLKIFSPNVLDITLVDL 155
E+ EKQ L F+ V DI L+++
Sbjct: 313 EDDTAFEKQSAL--FALAVSDIVLINM 387
>BI272296 similar to GP|12322014|gb GTPase putative; 34281-30152
{Arabidopsis thaliana}, partial (22%)
Length = 487
Score = 30.8 bits (68), Expect = 2.1
Identities = 12/25 (48%), Positives = 19/25 (76%)
Frame = +2
Query: 46 LPQVAVVGSQSSGKSSVLESLVGRD 70
+P +++VG + GKSS+L +LVG D
Sbjct: 173 VPAISIVGRPNVGKSSILNALVGED 247
>TC88009 similar to PIR|T48507|T48507 probable GTP-binding protein -
Arabidopsis thaliana, partial (72%)
Length = 1399
Score = 29.6 bits (65), Expect = 4.6
Identities = 17/45 (37%), Positives = 28/45 (61%)
Frame = +1
Query: 46 LPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVH 90
LP+ A+VG + GKSS+L S+V R L + +++P QL++
Sbjct: 622 LPEFALVGRSNVGKSSLLNSIVRRKKLA----LTSKKPGKTQLIN 744
>TC77596 similar to GP|18377702|gb|AAL67001.1 putative clathrin binding
protein {Arabidopsis thaliana}, partial (44%)
Length = 2019
Score = 29.6 bits (65), Expect = 4.6
Identities = 15/49 (30%), Positives = 28/49 (56%), Gaps = 4/49 (8%)
Frame = +3
Query: 350 LSKYCDAFSSMLEG----KNKEMSTCELSGGARIHYIFQSIYVRSLEAV 394
L K C S ML+ K++++ C++ R+H++FQ ++R+ E V
Sbjct: 996 LDKICSVTSWMLQHLFL*KSQQLVMCQMLISLRMHHLFQQHHMRTKELV 1142
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,747,714
Number of Sequences: 36976
Number of extensions: 256033
Number of successful extensions: 1294
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1270
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1281
length of query: 773
length of database: 9,014,727
effective HSP length: 104
effective length of query: 669
effective length of database: 5,169,223
effective search space: 3458210187
effective search space used: 3458210187
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0100c.1


