

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100a.5
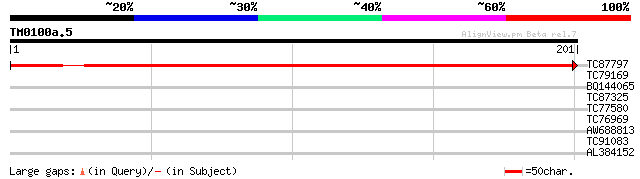
(201 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87797 similar to SP|O80813|Y20L_ARATH Ycf20-like protein. [Mou... 301 1e-82
TC79169 type IIB calcium ATPase [Medicago truncatula] 28 2.4
BQ144065 similar to PIR|T06291|T062 extensin homolog T9E8.80 - A... 28 2.4
TC87325 similar to GP|18176187|gb|AAL60000.1 unknown protein {Ar... 28 3.1
TC77580 similar to PIR|T10861|T10861 phaseolin G-box binding pro... 28 3.1
TC76969 similar to SP|P49231|PRO1_PHAVU Profilin 1. [Kidney bean... 27 5.3
AW688813 27 5.3
TC91083 similar to GP|1916643|gb|AAC49826.1| desacetoxyvindoline... 27 7.0
AL384152 26 9.1
>TC87797 similar to SP|O80813|Y20L_ARATH Ycf20-like protein. [Mouse-ear
cress] {Arabidopsis thaliana}, partial (89%)
Length = 1136
Score = 301 bits (771), Expect = 1e-82
Identities = 152/201 (75%), Positives = 167/201 (82%)
Frame = +2
Query: 1 MNLQWRLSITEHESSKKPYTAFSASRVHGCCCEPILSGLTSNAGGKSSHFPQSGFLGRKA 60
MNL +LSITE+E SK+ + +H CCC+P S LT N G K F QS FL R+
Sbjct: 212 MNLPSKLSITENEGSKRSF-------IHSCCCQPRFSALTPNVGMKPLPFLQSRFLRRRP 370
Query: 61 VWKIAFALNTGGVPGNGEQQSLNDTSSSFGGTRLGRILSAGGRQLLDKLNSARKNVPMKI 120
W +AFALNT G+ GNGEQQSLND+ SS GGTRLGRILSAGGRQLLDKLNSARKN P K+
Sbjct: 371 GWTVAFALNTDGLSGNGEQQSLNDSGSSLGGTRLGRILSAGGRQLLDKLNSARKNFPTKV 550
Query: 121 FLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLLYRKPPTVRTGRLQSFLLM 180
FLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGML+Y+KPPT RTGRLQSFL+M
Sbjct: 551 FLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLIYKKPPTTRTGRLQSFLVM 730
Query: 181 LNYWKAGICLGLFVDAFKLGS 201
+NYWKAGICLGLFVDAFKLGS
Sbjct: 731 VNYWKAGICLGLFVDAFKLGS 793
>TC79169 type IIB calcium ATPase [Medicago truncatula]
Length = 3565
Score = 28.1 bits (61), Expect = 2.4
Identities = 10/26 (38%), Positives = 16/26 (61%)
Frame = -1
Query: 32 CEPILSGLTSNAGGKSSHFPQSGFLG 57
C I+ +TSN+ + H PQ+G+ G
Sbjct: 2449 CRSIVGSITSNSYNLTKHIPQTGYQG 2372
>BQ144065 similar to PIR|T06291|T062 extensin homolog T9E8.80 - Arabidopsis
thaliana, partial (3%)
Length = 1397
Score = 28.1 bits (61), Expect = 2.4
Identities = 13/36 (36%), Positives = 20/36 (55%)
Frame = -3
Query: 157 GIGMLLYRKPPTVRTGRLQSFLLMLNYWKAGICLGL 192
GIG+ P ++ R S L ++YW G+C+GL
Sbjct: 504 GIGL------PYLQPSRRLSLLSCIHYWHPGLCIGL 415
>TC87325 similar to GP|18176187|gb|AAL60000.1 unknown protein {Arabidopsis
thaliana}, partial (50%)
Length = 1568
Score = 27.7 bits (60), Expect = 3.1
Identities = 18/43 (41%), Positives = 25/43 (57%), Gaps = 4/43 (9%)
Frame = +3
Query: 83 NDTSSSFG----GTRLGRILSAGGRQLLDKLNSARKNVPMKIF 121
N+T+SS G +L R +SAG D+ NSAR N+P +F
Sbjct: 492 NETNSSTGVISSDEQLARQMSAG----FDRQNSARHNMPAGLF 608
>TC77580 similar to PIR|T10861|T10861 phaseolin G-box binding protein PG1 -
kidney bean, partial (42%)
Length = 1383
Score = 27.7 bits (60), Expect = 3.1
Identities = 26/82 (31%), Positives = 37/82 (44%), Gaps = 4/82 (4%)
Frame = -1
Query: 96 RILSAGGRQLLDKLNSARKNVPMK----IFLLLLGFYTANALATILGQTGDWDVLVAGVV 151
R+ S+ + K ++RK V K FLL L + + LG W V+V VV
Sbjct: 381 RLYSSRHPNRVTKNLNSRKKVARK*RRSRFLLHLRLKPEDVVGECLG*IRWWFVVVVTVV 202
Query: 152 VAAIEGIGMLLYRKPPTVRTGR 173
V AI I KPP +++ R
Sbjct: 201 VVAIRLINQRRS*KPPLLKSYR 136
>TC76969 similar to SP|P49231|PRO1_PHAVU Profilin 1. [Kidney bean French
bean] {Phaseolus vulgaris}, complete
Length = 803
Score = 26.9 bits (58), Expect = 5.3
Identities = 14/23 (60%), Positives = 15/23 (64%)
Frame = +3
Query: 102 GRQLLDKLNSARKNVPMKIFLLL 124
G QLL L +KN P KIFLLL
Sbjct: 12 GTQLLLLLTQNKKNFPEKIFLLL 80
>AW688813
Length = 281
Score = 26.9 bits (58), Expect = 5.3
Identities = 16/48 (33%), Positives = 19/48 (39%)
Frame = -2
Query: 55 FLGRKAVWKIAFALNTGGVPGNGEQQSLNDTSSSFGGTRLGRILSAGG 102
F G K WK F L G G G+ S+ FGG + GG
Sbjct: 238 FFGXKTNWKTPFXLR--GKKGRGKVLXREGGSTPFGGXEISSGKPPGG 101
>TC91083 similar to GP|1916643|gb|AAC49826.1| desacetoxyvindoline
4-hydroxylase {Catharanthus roseus}, partial (30%)
Length = 606
Score = 26.6 bits (57), Expect = 7.0
Identities = 14/32 (43%), Positives = 15/32 (46%)
Frame = -3
Query: 20 TAFSASRVHGCCCEPILSGLTSNAGGKSSHFP 51
T + S V C PIL S GG S HFP
Sbjct: 256 TLLNLSFVISCRTSPILITRASGIGGTSIHFP 161
>AL384152
Length = 448
Score = 26.2 bits (56), Expect = 9.1
Identities = 17/64 (26%), Positives = 28/64 (43%)
Frame = -2
Query: 121 FLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLLYRKPPTVRTGRLQSFLLM 180
F+ L +Y +N + ++ D L + +EG+ + Y P GRL FL
Sbjct: 393 FIRYLAYYHSNIIFNVM-----CDCL***CLXKLLEGLNLSQYVPPQCSXLGRLPEFLEF 229
Query: 181 LNYW 184
L+ W
Sbjct: 228 LHCW 217
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,912,693
Number of Sequences: 36976
Number of extensions: 90711
Number of successful extensions: 460
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 458
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 459
length of query: 201
length of database: 9,014,727
effective HSP length: 91
effective length of query: 110
effective length of database: 5,649,911
effective search space: 621490210
effective search space used: 621490210
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0100a.5


