

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
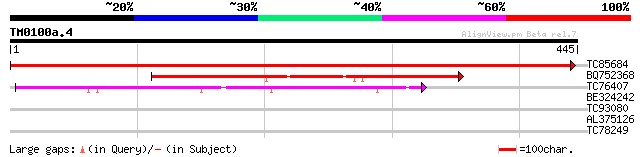
Query= TM0100a.4
(445 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85684 homologue to GP|3175990|emb|CAA06731.1 GDP dissociation ... 753 0.0
BQ752368 similar to PIR|T38215|T38 probable secretory pathway GD... 261 4e-70
TC76407 weakly similar to GP|20453227|gb|AAM19852.1 AT3g06540/F5... 112 3e-25
BE324242 weakly similar to GP|12322677|gb| Rab escort protein p... 32 0.38
TC93080 similar to PIR|T49718|T49718 probable hydroxymethylgluta... 30 2.4
AL375126 homologue to GP|3175990|emb| GDP dissociation inhibitor... 29 3.2
TC78249 similar to GP|16648993|gb|AAL24348.1 Unknown protein {Ar... 28 7.1
>TC85684 homologue to GP|3175990|emb|CAA06731.1 GDP dissociation inhibitor
{Cicer arietinum}, complete
Length = 1849
Score = 753 bits (1944), Expect = 0.0
Identities = 360/444 (81%), Positives = 404/444 (90%)
Frame = +2
Query: 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGASTSLNLTQLWKRFRGDDK 60
MDEEYDVIVLGTGLKEC+LSGLLSVDGLKVLHMDRNDYYGG STSLNL QLWK+FRGDDK
Sbjct: 137 MDEEYDVIVLGTGLKECVLSGLLSVDGLKVLHMDRNDYYGGESTSLNLIQLWKKFRGDDK 316
Query: 61 SPEHLGSSREFNVDMIPKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYNKGKIYKVPA 120
P HLGSS+++N+DM PKF+MANG LVRVLIHTDVTKYL FKAVDGSFV+NK K++KVP+
Sbjct: 317 PPPHLGSSKDYNIDMNPKFIMANGTLVRVLIHTDVTKYLYFKAVDGSFVFNKAKVHKVPS 496
Query: 121 TDVEALKSPLMGLFEKRRARKFFIYVQDYEVSDPKSHEGLDLNQVTARQLISKYGLEDDT 180
D+EALKSPLMG+FEKRRARKFFIYVQDY SDPK+H+G+DL +VT ++LI+K+GL+D+T
Sbjct: 497 NDMEALKSPLMGIFEKRRARKFFIYVQDYNESDPKTHDGMDLTRVTTKELIAKFGLDDNT 676
Query: 181 IDFIGHALALHLDDSYLDTPAKDFVERIKVYAESLARFQGGSPYIYPLYGLGELPQAFAR 240
+DFIGHALALH DD YL+ PA D V+R+K+YAESLARFQGGSPYIYPLYGLGELPQAFAR
Sbjct: 677 VDFIGHALALHRDDRYLNEPALDTVKRMKLYAESLARFQGGSPYIYPLYGLGELPQAFAR 856
Query: 241 LSAVYGGTYMLNKPECKVEFDSAGKAIGVTSEGETAKCKKVVCDPSYLPDKVRNVGKVAR 300
LSAVYGGTYMLNKPECKVEFD GK IGVTSEGETAKCKK+VCDPSYL +KVR VGKVAR
Sbjct: 857 LSAVYGGTYMLNKPECKVEFDDQGKVIGVTSEGETAKCKKLVCDPSYLTNKVRKVGKVAR 1036
Query: 301 AICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHNVAPKGKYIAFVTTEAET 360
AI IMSHPIP+TNDS S QVILPQKQLGRKSDMYLFCCSY+HNV PKGK+IAFV+TEAET
Sbjct: 1037AIAIMSHPIPNTNDSQSVQVILPQKQLGRKSDMYLFCCSYSHNVCPKGKFIAFVSTEAET 1216
Query: 361 DQPEVELKPGIDLLGPVDETFYDIYDRYEPTNDHAADGCFISKSYDATTHFETTVKDVVE 420
DQP +ELKPGIDLLGPVDE F+D+YDR+EP N+ D CFIS SYDATTHFE+TV DV+
Sbjct: 1217DQPAIELKPGIDLLGPVDEIFFDMYDRFEPVNEPTLDNCFISTSYDATTHFESTVLDVLN 1396
Query: 421 IYSKITGKVLDLSVDLSAASATAE 444
+Y+ ITGKVLDL+VDLSAASA E
Sbjct: 1397MYTLITGKVLDLTVDLSAASAAEE 1468
>BQ752368 similar to PIR|T38215|T38 probable secretory pathway GDP
dissociation inhibitor - fission yeast
(Schizosaccharomyces pombe), partial (44%)
Length = 811
Score = 261 bits (667), Expect = 4e-70
Identities = 134/254 (52%), Positives = 180/254 (70%), Gaps = 9/254 (3%)
Frame = +2
Query: 112 KGKIYKVPATDVEALKSPLMGLFEKRRARKFFIYVQDYEVSDPKSHEGLDLNQVTARQLI 171
K + KVP+ EAL+SPLMG+FEKRR + F ++ +E D +H+GLD+N VT +++
Sbjct: 11 KATVAKVPSDAGEALRSPLMGIFEKRRMKSFIEWIGSFERKDTATHKGLDINNVTMKEVY 190
Query: 172 SKYGLEDDTIDFIGHALALHLDDSYLDTP--AKDFVERIKVYAESLARFQGGSPYIYPLY 229
K+GLE T DFIGHA+AL D Y++ P A + +ERI++Y S+AR+ G SPYIYPLY
Sbjct: 191 DKFGLETGTRDFIGHAMALFPTDDYINKPGAAPEAIERIRLYGTSVARY-GKSPYIYPLY 367
Query: 230 GLGELPQAFARLSAVYGGTYMLNKPECKVEFDSAGKAIGV--TSEGET-----AKCKKVV 282
GLGELPQ FARLSA+YGGTYML+ +V ++ GKA+G+ T G K K ++
Sbjct: 368 GLGELPQGFARLSAIYGGTYMLHTNVDEVTYEG-GKAVGIKATMSGVEEMKFETKAKMII 544
Query: 283 CDPSYLPDKVRNVGKVARAICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAH 342
DPSY P KV+ VG+V RAICI+ HP+ TNDS S+Q+I+PQ Q+GRK+D+Y+ C S AH
Sbjct: 545 GDPSYFPGKVKVVGQVLRAICILKHPLAGTNDSDSSQLIIPQSQIGRKNDIYIACVSSAH 724
Query: 343 NVAPKGKYIAFVTT 356
NV PKG +IA V+T
Sbjct: 725 NVCPKGYWIAIVST 766
>TC76407 weakly similar to GP|20453227|gb|AAM19852.1 AT3g06540/F5E6_13
{Arabidopsis thaliana}, partial (62%)
Length = 2233
Score = 112 bits (280), Expect = 3e-25
Identities = 105/392 (26%), Positives = 162/392 (40%), Gaps = 69/392 (17%)
Frame = +3
Query: 5 YDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGASTSLNLTQLWKRFRGDDK---- 60
+D+I++GTGL E I+S S G +LH+D N +YG SL+ L K
Sbjct: 126 FDLIIVGTGLSESIISAAASAVGKTILHLDPNPFYGSHFASLSFEDLISHLNSPPKYITA 305
Query: 61 ----------------------SPEHLGS-------------SREFNVDMI-PKFMMANG 84
S S SR+FN+D+ P+ + +
Sbjct: 306 TATATTDNSDYTTVNLIQQTIFSDAEFNSCNSIEESQFLRENSRKFNLDLAGPRALFSAD 485
Query: 85 ALVRVLIHTDVTKYLNFKAVDGSFVYNKGK-IYKVPATDVEALKSPLMGLFEKRRARKFF 143
+ +L+ + +YL FK +D S VYN + + VP + + + L EK + KFF
Sbjct: 486 KTIDLLLKSGAAQYLEFKGIDASLVYNTNEGLVNVPDSRGAIFRDKNLSLKEKNQLMKFF 665
Query: 144 IYVQDY---EVSDPKSHEGLDLNQVTARQLISKYGLEDDTIDFIGHALALHLDDSYLDTP 200
VQ + + S E ++ + V+ + K GL + +A+A+ D
Sbjct: 666 KLVQQHLGGNEDEKISEEDMESSFVS---YLEKLGLPSKIKAILLYAIAMVDFDQENGGV 836
Query: 201 AKDF------VERIKVYAESLARFQGG-SPYIYPLYGLGELPQAFARLSAVYGGTYMLNK 253
KD ++R+ Y+ S+ R+ +YP+YG GELPQAF R +AV G Y+L
Sbjct: 837 CKDLLKTKDGIDRLAQYSSSVGRYPNAPGALLYPIYGEGELPQAFCRRAAVKGCIYVLRM 1016
Query: 254 PECKVEFDS-AGKAIGV-TSEGETAKCKKVVCDPSY----------------LPDKVRNV 295
P + D G GV S G+ K++ DPS+ + V
Sbjct: 1017PVISLLIDKVTGSYKGVRLSSGQDLYSHKLILDPSFTIPSTPSLSPKDFSLQMLSHVDIK 1196
Query: 296 GKVARAICIMSHPIPDTNDSHSAQVILPQKQL 327
G VAR ICI I D + V+ P + L
Sbjct: 1197GMVARGICITRSSIKP--DVSNCSVVYPPRSL 1286
>BE324242 weakly similar to GP|12322677|gb| Rab escort protein putative;
48705-51166 {Arabidopsis thaliana}, partial (10%)
Length = 478
Score = 32.3 bits (72), Expect = 0.38
Identities = 15/46 (32%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Frame = +1
Query: 67 SSREFNVDMI-PKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYN 111
+SR+FN+D+ P+ + + + +L++ + NFK +D S VYN
Sbjct: 295 NSRKFNLDLAGPRALFSADKTIDLLLNQVLHSTYNFKGIDASLVYN 432
>TC93080 similar to PIR|T49718|T49718 probable hydroxymethylglutaryl-CoA
synthase [imported] - Neurospora crassa, partial (33%)
Length = 662
Score = 29.6 bits (65), Expect = 2.4
Identities = 16/60 (26%), Positives = 29/60 (47%)
Frame = +3
Query: 386 DRYEPTNDHAADGCFISKSYDATTHFETTVKDVVEIYSKITGKVLDLSVDLSAASATAEE 445
DR++ HA +SKSY + + I++++ ++ DLS D S + T E+
Sbjct: 150 DRFDYMAFHAPTCKLVSKSYARMLYNDYVQNPSNPIFAEVPAEIKDLSYDASVSDRTVEK 329
>AL375126 homologue to GP|3175990|emb| GDP dissociation inhibitor {Cicer
arietinum}, partial (6%)
Length = 404
Score = 29.3 bits (64), Expect = 3.2
Identities = 14/15 (93%), Positives = 15/15 (99%)
Frame = -3
Query: 427 GKVLDLSVDLSAASA 441
GKVLDL+VDLSAASA
Sbjct: 288 GKVLDLTVDLSAASA 244
>TC78249 similar to GP|16648993|gb|AAL24348.1 Unknown protein {Arabidopsis
thaliana}, partial (55%)
Length = 1095
Score = 28.1 bits (61), Expect = 7.1
Identities = 21/66 (31%), Positives = 34/66 (50%), Gaps = 4/66 (6%)
Frame = +1
Query: 19 LSGLLSVDGLKVLHMDRNDYYGGASTSLNLTQLWKRFR--GDDKSPEHLGSSREF--NVD 74
+SGL S GL ++H + + + T+LNL KRF + L SSR F +++
Sbjct: 43 VSGLRSSCGLPLVHHHNHLRFSSSFTNLNLHSKSKRFTLFARYAQTQDLFSSRRFQDSIE 222
Query: 75 MIPKFM 80
+PK +
Sbjct: 223 KLPKLV 240
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,240,648
Number of Sequences: 36976
Number of extensions: 159797
Number of successful extensions: 631
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 625
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 629
length of query: 445
length of database: 9,014,727
effective HSP length: 99
effective length of query: 346
effective length of database: 5,354,103
effective search space: 1852519638
effective search space used: 1852519638
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0100a.4


