

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
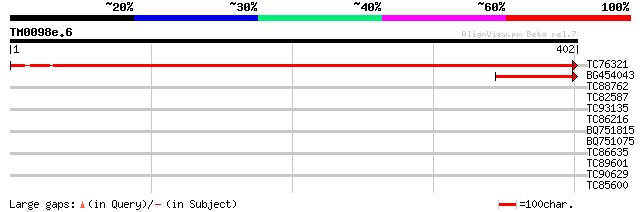
Query= TM0098e.6
(402 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76321 homologue to GP|16033631|gb|AAL13304.1 leucine zipper-co... 727 0.0
BG454043 similar to GP|2213425|emb| hypothetical protein {Citrus... 77 2e-14
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 30 1.3
TC82587 similar to PIR|H86313|H86313 protein F2H15.10 [imported]... 30 1.3
TC93135 weakly similar to GP|21327037|gb|AAM48133.1 putative fla... 30 1.7
TC86216 similar to GP|15982915|gb|AAL09804.1 AT4g01050/F2N1_31 {... 29 3.7
BQ751815 similar to PIR|S58652|S586 hypothetical protein YFR036w... 29 3.7
BQ751075 similar to SP|P11913|MPPB Mitochondrial processing pept... 28 8.2
TC86635 weakly similar to GP|9758600|dbj|BAB09233.1 gene_id:MDF2... 28 8.2
TC89601 similar to GP|9652289|gb|AAF91469.1| putative vacuolar p... 28 8.2
TC90629 similar to GP|15984218|emb|CAC93760. unnamed protein pro... 28 8.2
TC85600 homologue to SP|P29828|PDI_MEDSA Protein disulfide isome... 28 8.2
>TC76321 homologue to GP|16033631|gb|AAL13304.1 leucine zipper-containing
protein {Euphorbia esula}, partial (92%)
Length = 1468
Score = 727 bits (1877), Expect = 0.0
Identities = 366/403 (90%), Positives = 375/403 (92%), Gaps = 1/403 (0%)
Frame = +2
Query: 1 MALVKPISKFTNAAPKFSNPRTISHSKFSTVRMSAT-TTPPSSTKPSKKANKTSIKETLL 59
MALVKPISKFT PKFSN + S +S +RMSAT T P+STKPSKK NKT+IKETLL
Sbjct: 65 MALVKPISKFT---PKFSNIPRRTVSSYS-IRMSATPTATPTSTKPSKKPNKTAIKETLL 232
Query: 60 TPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDG 119
TPRFYTTDFDEMETLFNTEINKNLNE EFEALLQEFKTDYNQTHFVRNKEFKEAADKLDG
Sbjct: 233 TPRFYTTDFDEMETLFNTEINKNLNEDEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDG 412
Query: 120 PLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGL 179
PLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGL
Sbjct: 413 PLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGL 592
Query: 180 SDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQCYPIF 239
SDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLK NPEYQCYPIF
Sbjct: 593 SDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKQNPEYQCYPIF 772
Query: 240 KYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYE 299
KYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYE
Sbjct: 773 KYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYE 952
Query: 300 GIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDIP 359
GIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR+LDRMV IN+KILA+GE+DD
Sbjct: 953 GIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRKLDRMVVINQKILAIGETDDNS 1132
Query: 360 FVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFEPQLVY 402
VKN KRIPLIAAL SELLA YLM PIESGSVD EFE LVY
Sbjct: 1133VVKNFKRIPLIAALVSELLAAYLMKPIESGSVDFPEFERGLVY 1261
>BG454043 similar to GP|2213425|emb| hypothetical protein {Citrus x
paradisi}, partial (18%)
Length = 663
Score = 76.6 bits (187), Expect = 2e-14
Identities = 41/58 (70%), Positives = 43/58 (73%)
Frame = +3
Query: 345 INEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFEPQLVY 402
IN ILA+GE+DD VKN RIPLIAAL SELLA YLM PIESGSV EFE LVY
Sbjct: 351 INXXILAIGETDDNSVVKNFXRIPLIAALVSELLAAYLMKPIESGSVXFPEFERGLVY 524
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 30.4 bits (67), Expect = 1.3
Identities = 14/38 (36%), Positives = 24/38 (62%)
Frame = +3
Query: 16 KFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTS 53
+F+NP + S S ++ +S TT+PP+ST S + T+
Sbjct: 72 QFNNPTSPSSSPSASSPVSTTTSPPASTPASSPVSTTT 185
>TC82587 similar to PIR|H86313|H86313 protein F2H15.10 [imported] -
Arabidopsis thaliana, partial (24%)
Length = 764
Score = 30.4 bits (67), Expect = 1.3
Identities = 47/185 (25%), Positives = 70/185 (37%), Gaps = 2/185 (1%)
Frame = +2
Query: 18 SNPRTISHSKFSTVR-MSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLFN 76
+ P+T S FS+++ + T + P S P K+ N ++ +
Sbjct: 188 NKPKTTSFKLFSSLKDENETNSSPVSIAPKKQDNISNNNN-------------------D 310
Query: 77 TEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAE 136
++ + NE E E ++KTD F+ N EAA KL E+ T
Sbjct: 311 DDVGRETNEDEKEQQEMDWKTDEEFKKFMGNPSI-EAAIKL------------EKKRTDR 451
Query: 137 FSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARK 196
KEL K NP+V +L+ R+ L K F ALDL L
Sbjct: 452 -----KLKELDTESSKNNPIVGVFNNLVRRNLILEKERLEKVEETFK-ALDLNKLKSCFG 613
Query: 197 Y-TFF 200
+ TFF
Sbjct: 614 FDTFF 628
>TC93135 weakly similar to GP|21327037|gb|AAM48133.1 putative flavanone
3-hydroxylase {Saussurea medusa}, partial (32%)
Length = 645
Score = 30.0 bits (66), Expect = 1.7
Identities = 18/63 (28%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Frame = +2
Query: 210 YLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDENRH----GDFFSALMKAQPQ 265
Y SEK+ WR N +QCYP+ ++ W ++ R+ GDF + + K +
Sbjct: 416 YDSEKVHLWR---------DNLRHQCYPLEQWQHIWPENPPRYRECVGDFSTEVKKLGSR 568
Query: 266 FLN 268
+N
Sbjct: 569 IMN 577
>TC86216 similar to GP|15982915|gb|AAL09804.1 AT4g01050/F2N1_31 {Arabidopsis
thaliana}, partial (54%)
Length = 1877
Score = 28.9 bits (63), Expect = 3.7
Identities = 22/79 (27%), Positives = 32/79 (39%), Gaps = 9/79 (11%)
Frame = +1
Query: 19 NPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTP---------RFYTTDFD 69
N T+S SKF +S+ P + ++ T+LTP + T F
Sbjct: 46 NIPTLSDSKFHDSSLSSKL*EPHKVSVVQNYAMEALNSTILTPLSVLSDRTKHKHPTKFS 225
Query: 70 EMETLFNTEINKNLNEHEF 88
+ F+T INK N EF
Sbjct: 226 CKASNFSTSINKKQNLREF 282
>BQ751815 similar to PIR|S58652|S586 hypothetical protein YFR036w-a - yeast
(Saccharomyces cerevisiae), partial (9%)
Length = 719
Score = 28.9 bits (63), Expect = 3.7
Identities = 14/38 (36%), Positives = 18/38 (46%)
Frame = +1
Query: 24 SHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTP 61
S + FS+ PPSST+P K NK+ TP
Sbjct: 199 SRTSFSSTTRPEPCKPPSSTRPPPKGNKSPANYRAPTP 312
>BQ751075 similar to SP|P11913|MPPB Mitochondrial processing peptidase beta
subunit mitochondrial precursor (EC 3.4.24.64)
(Beta-MPP), partial (23%)
Length = 344
Score = 27.7 bits (60), Expect = 8.2
Identities = 13/50 (26%), Positives = 27/50 (54%)
Frame = +2
Query: 12 NAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTP 61
+A ++PR + S+++ + + T PPSST PS + ++ + +P
Sbjct: 56 SATSSSASPRRL-RSRWTRLSSTTFTPPPSSTSPSAAPSSAPVRTSATSP 202
>TC86635 weakly similar to GP|9758600|dbj|BAB09233.1
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1963
Score = 27.7 bits (60), Expect = 8.2
Identities = 11/40 (27%), Positives = 22/40 (54%)
Frame = -3
Query: 203 KFIFYATYLSEKIGYWRYITIYRHLKANPEYQCYPIFKYF 242
+F F+ LS ++ +WR + + HL+ + + Y F+ F
Sbjct: 1109 QFCFFRRTLSLQMMFWRVFSFFSHLQIHSQNHSYLPFQIF 990
>TC89601 similar to GP|9652289|gb|AAF91469.1| putative vacuolar proton
ATPase subunit E {Lycopersicon esculentum}, partial
(52%)
Length = 832
Score = 27.7 bits (60), Expect = 8.2
Identities = 10/29 (34%), Positives = 16/29 (54%)
Frame = -2
Query: 224 YRHLKANPEYQCYPIFKYFENWCQDENRH 252
+ H K P +Q + +Y + W Q E+RH
Sbjct: 366 FHHEKPTPPHQSSKVREYCDAWVQGEDRH 280
>TC90629 similar to GP|15984218|emb|CAC93760. unnamed protein product
{Lycopersicon esculentum}, partial (24%)
Length = 745
Score = 27.7 bits (60), Expect = 8.2
Identities = 18/50 (36%), Positives = 25/50 (50%), Gaps = 4/50 (8%)
Frame = +1
Query: 24 SHSKFSTVRMSAT----TTPPSSTKPSKKANKTSIKETLLTPRFYTTDFD 69
SHS ST+ +AT +P SS K +KK TS++ TDF+
Sbjct: 178 SHSNSSTIFCTATKSQKNSPSSSKKKTKKKKNTSVESDDWKNLNSNTDFE 327
>TC85600 homologue to SP|P29828|PDI_MEDSA Protein disulfide isomerase
precursor (PDI) (EC 5.3.4.1). [Alfalfa] {Medicago
sativa}, complete
Length = 1915
Score = 27.7 bits (60), Expect = 8.2
Identities = 13/45 (28%), Positives = 24/45 (52%)
Frame = +2
Query: 88 FEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERS 132
F AL ++ ++DY+ H + K + + GP+ ++F F E S
Sbjct: 593 FIALAEKLRSDYDFGHTLNAKHLPKGDSSVSGPVVRLFKPFDELS 727
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,868,813
Number of Sequences: 36976
Number of extensions: 171261
Number of successful extensions: 1063
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1057
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1059
length of query: 402
length of database: 9,014,727
effective HSP length: 98
effective length of query: 304
effective length of database: 5,391,079
effective search space: 1638888016
effective search space used: 1638888016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0098e.6


