

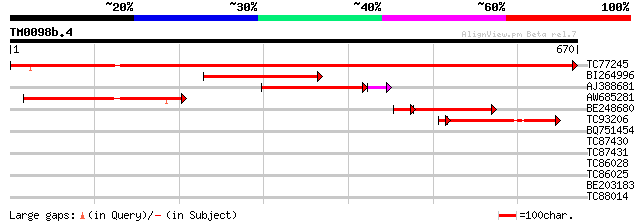
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098b.4
(670 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77245 similar to GP|6630556|gb|AAF19575.1| putative alpha-L-ar... 1155 0.0
BI264996 weakly similar to GP|16417958|gb arabinosidase ARA-1 {L... 228 6e-60
AJ388681 similar to GP|13398412|gb arabinoxylan arabinofuranohyd... 212 2e-56
AW685281 weakly similar to GP|17380938|gb putative arabinosidase... 201 1e-51
BE248680 weakly similar to GP|13937191|gb AT3g10740/T7M13_18 {Ar... 134 7e-37
TC93206 similar to GP|13398412|gb|AAK21879.1 arabinoxylan arabin... 126 7e-31
BQ751454 weakly similar to GP|23326899|gb| alpha-L-arabinosidase... 34 0.16
TC87430 34 0.21
TC87431 34 0.21
TC86028 similar to GP|7677262|gb|AAF67098.1| delta-COP {Zea mays... 32 1.0
TC86025 similar to GP|7677262|gb|AAF67098.1| delta-COP {Zea mays... 31 1.3
BE203183 30 2.3
TC88014 similar to GP|15215786|gb|AAK91438.1 AT3g43520/T18D12_90... 29 6.7
>TC77245 similar to GP|6630556|gb|AAF19575.1| putative
alpha-L-arabinofuranosidase {Arabidopsis thaliana},
partial (89%)
Length = 2351
Score = 1155 bits (2989), Expect = 0.0
Identities = 566/676 (83%), Positives = 611/676 (89%), Gaps = 6/676 (0%)
Frame = +2
Query: 1 MGLYKASFIVAALQLCIAVSLCA----IKVNADLNATLTVDAHETSGKPISETLFGLFFE 56
MG K S V LQL I V L ++V ADLNATL VDA + SG+ I ETLFG+FFE
Sbjct: 203 MGFSKDSCCVFMLQLLIVVYLVVQCFDVQVQADLNATLVVDASQASGRRIPETLFGIFFE 382
Query: 57 EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIAL 116
EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGN ++INVETDRTSCF+RNK+AL
Sbjct: 383 EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATYINVETDRTSCFERNKVAL 562
Query: 117 RLEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNG 176
RLEVLCD CP DGVGV+NPGFWGMNIEQGKKYKVVFY RSTG L+L VSLTGSNG
Sbjct: 563 RLEVLCDGT----CPTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNLKVSLTGSNG 730
Query: 177 V--LASNVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTY 234
V LAS VITGSASDFSNW KVET+LEAKATN NSRLQLTTT KGVIWLDQVSAMPLDTY
Sbjct: 731 VGSLASTVITGSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKGVIWLDQVSAMPLDTY 910
Query: 235 KGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKY 294
KG GFRS+L+ ML DLKP FIRFPGGCFVEG++LRNAFRWK ++GPWEERPGHFGDVWKY
Sbjct: 911 KGHGFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKY 1090
Query: 295 WTDDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPT 354
WTDDGLGY+EFLQL+EDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDG+EFARGDPT
Sbjct: 1091WTDDGLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPT 1270
Query: 355 SKWGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGS 414
SKWGS+RA+MGHPEPFNLKYVAVGNEDCGKKNYRGNYL+FY AIR AYPDIQIISNCDGS
Sbjct: 1271SKWGSMRAAMGHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIISNCDGS 1450
Query: 415 SRPLDHPADIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAI 474
SRPLDHPAD+YDYHIYTNANDMFSRS+TFN RSGPKAFVSEYAVTGNDAG GSLLAA+
Sbjct: 1451SRPLDHPADMYDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAAL 1630
Query: 475 AEAGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSES 534
AEAGFL+GLEKNSDIV M SYAPLFVNANDRRWNPDAIVFNSFQ YGTPSYW+QLFFSES
Sbjct: 1631AEAGFLIGLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSES 1810
Query: 535 SGATLLNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNS 594
+GATLLNSSLQT++S+SL+ASAI WQ+S DKKNYIRIK VNFG + VNLKIS NGLDPNS
Sbjct: 1811NGATLLNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNS 1990
Query: 595 LQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKES 654
LQ SGST+TVLTS N+MDENSFSQPKKV+PIQSLLQ+VGK+MNV+VPPHSF+SFDLLKES
Sbjct: 1991LQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLLKES 2170
Query: 655 SNLKMVGGGSSTRSSI 670
SNLKM+ SS+ SSI
Sbjct: 2171SNLKMLESDSSSWSSI 2218
>BI264996 weakly similar to GP|16417958|gb arabinosidase ARA-1 {Lycopersicon
esculentum}, partial (20%)
Length = 422
Score = 228 bits (581), Expect = 6e-60
Identities = 100/140 (71%), Positives = 117/140 (83%)
Frame = +2
Query: 230 PLDTYKGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFG 289
PLDTYKG GFR L M+A+LKPR RFPGGC+VEG L+NAF+WK++IGPWE RPGH+G
Sbjct: 2 PLDTYKGHGFRMNLFQMVAELKPRXFRFPGGCYVEGNVLKNAFQWKQTIGPWENRPGHYG 181
Query: 290 DVWKYWTDDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFA 349
DVW YWTDDG G+FE LQLAEDL ALPIWVFNNG+SH+++V+ SA+ P VQ ALDGIE A
Sbjct: 182 DVWDYWTDDGFGFFEGLQLAEDLNALPIWVFNNGISHSEQVNVSAISPSVQXALDGIEXA 361
Query: 350 RGDPTSKWGSIRASMGHPEP 369
G PTS+WGSI A MGHP+P
Sbjct: 362 IGSPTSRWGSIIAXMGHPKP 421
>AJ388681 similar to GP|13398412|gb arabinoxylan arabinofuranohydrolase
isoenzyme AXAH-I {Hordeum vulgare}, partial (18%)
Length = 574
Score = 212 bits (540), Expect(2) = 2e-56
Identities = 96/125 (76%), Positives = 109/125 (86%)
Frame = +1
Query: 298 DGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKW 357
DGLG+FE LQLAED+GALPIWVFNNG+SH+DEVDTS + PFV+EAL+GIEFARG TSKW
Sbjct: 1 DGLGFFEGLQLAEDIGALPIWVFNNGISHSDEVDTSVISPFVKEALEGIEFARGSSTSKW 180
Query: 358 GSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRP 417
GS+RASMGHP+PFNLKYVA+GNEDC KKNY GNY+ FY AI+ YPDIQIISNC P
Sbjct: 181 GSVRASMGHPKPFNLKYVAIGNEDCYKKNYYGNYMAFYKAIKKFYPDIQIISNCPAFKTP 360
Query: 418 LDHPA 422
L+HPA
Sbjct: 361 LNHPA 375
Score = 25.8 bits (55), Expect(2) = 2e-56
Identities = 12/30 (40%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Frame = +2
Query: 423 DIYDYHIY-TNANDMFSRSSTFNTALRSGP 451
D+YDYH Y +A MF+ F+ + GP
Sbjct: 377 DLYDYHTYPIDARXMFNAYHEFDKSPXHGP 466
>AW685281 weakly similar to GP|17380938|gb putative arabinosidase
{Arabidopsis thaliana}, partial (18%)
Length = 651
Score = 201 bits (510), Expect = 1e-51
Identities = 102/198 (51%), Positives = 138/198 (69%), Gaps = 5/198 (2%)
Frame = +2
Query: 17 IAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVSNRGF 76
I VS A + NA+ + ++ + G+P+ TLFG+F+EEINHAG+GG+WA+LV+N GF
Sbjct: 68 IIVSFVAFQCNANGSQISSLVVNAAQGRPMPNTLFGIFYEEINHAGSGGIWAQLVNNSGF 247
Query: 77 EAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPADGVG 136
EA G PSNI PW+IIG ES + ++T+ +SCF+RNK+ALR++VLCD CP DGVG
Sbjct: 248 EAAGTRTPSNIFPWTIIGTESSVKLQTELSSCFERNKVALRMDVLCDK-----CPPDGVG 412
Query: 137 VFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGS--NGVLASNVI---TGSASDFS 191
V NPGFWGMNI QGKKYKVVF+ RS GSLD+ V+ + +LAS+ I S
Sbjct: 413 VSNPGFWGMNIVQGKKYKVVFFYRSLGSLDMRVAFRDAIXGRILASSHIIRHKASKKRVP 592
Query: 192 NWKKVETLLEAKATNHNS 209
+ V+T+LEA+A++ NS
Sbjct: 593 KXQXVQTILEARASSXNS 646
>BE248680 weakly similar to GP|13937191|gb AT3g10740/T7M13_18 {Arabidopsis
thaliana}, partial (15%)
Length = 375
Score = 134 bits (338), Expect(2) = 7e-37
Identities = 66/98 (67%), Positives = 78/98 (79%)
Frame = +1
Query: 478 GFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGA 537
GFL+GLEKNSD+V MV+YAPLFVN NDR+W PDAIVF+S Q YG PSYW+ F ESSGA
Sbjct: 76 GFLIGLEKNSDVVSMVNYAPLFVNTNDRKWTPDAIVFDSHQVYGIPSYWLIKLFKESSGA 255
Query: 538 TLLNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVN 575
T LNS+LQT S +L ASAI+W+SS D + +RIKV N
Sbjct: 256 TFLNSTLQT-DSPTLAASAISWKSSVDGTSILRIKVAN 366
Score = 38.1 bits (87), Expect(2) = 7e-37
Identities = 18/26 (69%), Positives = 21/26 (80%)
Frame = +3
Query: 454 FVSEYAVTGNDAGTGSLLAAIAEAGF 479
FVSEYA+ DAG G+LLAA+AEA F
Sbjct: 3 FVSEYALIKEDAGNGTLLAAVAEARF 80
>TC93206 similar to GP|13398412|gb|AAK21879.1 arabinoxylan
arabinofuranohydrolase isoenzyme AXAH-I {Hordeum
vulgare}, partial (6%)
Length = 642
Score = 126 bits (316), Expect(2) = 7e-31
Identities = 67/133 (50%), Positives = 88/133 (65%)
Frame = +1
Query: 518 QSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVNFG 577
Q YGTPSYWV F ES+GAT LNS+LQT+ +L ASAI + +K Y++IK+ N
Sbjct: 40 QVYGTPSYWVTYLFKESNGATFLNSTLQTTDPGTLAASAILVKDPQNKNTYLKIKIANMR 219
Query: 578 ANTVNLKISLNGLDPNSLQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMN 637
V+ KIS+ G +L+ GST+TVLT N +DENSF++PKK+ P S LQN G EMN
Sbjct: 220 KTQVDFKISIQGFASKNLK--GSTKTVLT-GNELDENSFAEPKKIAPQTSPLQNPGNEMN 390
Query: 638 VVVPPHSFSSFDL 650
V+V P S + D+
Sbjct: 391 VIVQPTSLTILDM 429
Score = 26.6 bits (57), Expect(2) = 7e-31
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = +2
Query: 507 WNPDAIVFNSFQSYG 521
WNPDAIVF+S + G
Sbjct: 8 WNPDAIVFSSIKFMG 52
>BQ751454 weakly similar to GP|23326899|gb| alpha-L-arabinosidase
{Bifidobacterium longum NCC2705}, partial (12%)
Length = 780
Score = 34.3 bits (77), Expect = 0.16
Identities = 16/48 (33%), Positives = 26/48 (53%)
Frame = +3
Query: 238 GFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERP 285
GFR ++V + +L +R+PGG F + W + +GP E+RP
Sbjct: 255 GFRKDVVEAVKELNCPVMRYPGGNFCA------TYHWIDGVGPREKRP 380
>TC87430
Length = 649
Score = 33.9 bits (76), Expect = 0.21
Identities = 12/21 (57%), Positives = 16/21 (76%)
Frame = +3
Query: 259 GGCFVEGEWLRNAFRWKESIG 279
GGCFVE +LRN +WK ++G
Sbjct: 459 GGCFVERNYLRNEIQWKAAVG 521
>TC87431
Length = 662
Score = 33.9 bits (76), Expect = 0.21
Identities = 12/21 (57%), Positives = 16/21 (76%)
Frame = +2
Query: 259 GGCFVEGEWLRNAFRWKESIG 279
GGCFVE +LRN +WK ++G
Sbjct: 437 GGCFVERNYLRNEIQWKAAVG 499
>TC86028 similar to GP|7677262|gb|AAF67098.1| delta-COP {Zea mays}, partial
(34%)
Length = 992
Score = 31.6 bits (70), Expect = 1.0
Identities = 14/53 (26%), Positives = 29/53 (54%), Gaps = 1/53 (1%)
Frame = +3
Query: 83 IPSNIDPW-SIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPADG 134
+P I+ W S GNE+++N+E + +S FD + + + + + ++ DG
Sbjct: 204 VPLTINCWPSSSGNETYVNIEYEASSMFDLRNVVISVPLPALREAPSVSQIDG 362
>TC86025 similar to GP|7677262|gb|AAF67098.1| delta-COP {Zea mays}, partial
(90%)
Length = 1890
Score = 31.2 bits (69), Expect = 1.3
Identities = 14/53 (26%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Frame = +1
Query: 83 IPSNIDPW-SIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPADG 134
+P I+ W S GNE+++N+E + +S FD + + + + ++ DG
Sbjct: 1264 VPLTINCWPSSAGNETYVNIEYEASSMFDLRNVVISVPFPALREAPSVSQIDG 1422
>BE203183
Length = 408
Score = 30.4 bits (67), Expect = 2.3
Identities = 11/17 (64%), Positives = 13/17 (75%)
Frame = +2
Query: 259 GGCFVEGEWLRNAFRWK 275
GGCFVE +LRN +WK
Sbjct: 356 GGCFVERNYLRNEIQWK 406
>TC88014 similar to GP|15215786|gb|AAK91438.1 AT3g43520/T18D12_90
{Arabidopsis thaliana}, partial (44%)
Length = 1052
Score = 28.9 bits (63), Expect = 6.7
Identities = 14/56 (25%), Positives = 26/56 (46%)
Frame = -3
Query: 366 HPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLDHP 421
+P P ++ + G + ++ +R K A++ P ++ S C S PL HP
Sbjct: 627 NPSPQEVQQICQGLQMMQEQVFRVAR*KHKRAVQKTNPQPKVTSGCHSSESPLHHP 460
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,265,720
Number of Sequences: 36976
Number of extensions: 309576
Number of successful extensions: 1418
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1399
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1410
length of query: 670
length of database: 9,014,727
effective HSP length: 103
effective length of query: 567
effective length of database: 5,206,199
effective search space: 2951914833
effective search space used: 2951914833
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0098b.4


