

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098b.1
(696 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
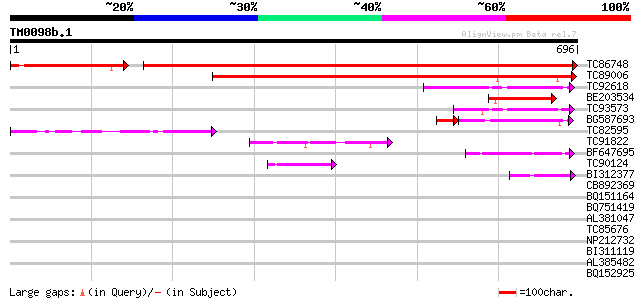
Score E
Sequences producing significant alignments: (bits) Value
TC86748 similar to GP|15485584|emb|CAC67503. SET-domain-containi... 926 0.0
TC89006 weakly similar to GP|20466308|gb|AAM20471.1 unknown prot... 534 e-152
TC92618 weakly similar to PIR|T02416|T02416 probable SET-domain ... 115 6e-26
BE203534 similar to GP|10178033|dbj SET-domain protein-like {Ara... 108 7e-24
TC93573 weakly similar to PIR|T02416|T02416 probable SET-domain ... 84 2e-16
BG587693 weakly similar to GP|17066863|gb Su(VAR)3-9-related pro... 64 7e-15
TC82595 similar to GP|10178033|dbj|BAB11516. SET-domain protein-... 78 1e-14
TC91822 similar to PIR|E96612|E96612 probable transcription fact... 63 3e-10
BF647695 similar to GP|6006866|gb| hypothetical protein {Arabido... 49 7e-06
TC90124 similar to GP|17529304|gb|AAL38879.1 putative transcript... 45 9e-05
BI312377 similar to GP|8843772|dbj contains similarity to zinc f... 45 9e-05
CB892369 weakly similar to GP|18376303|em related to regulatory ... 42 0.001
BQ151164 40 0.003
BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania maj... 34 0.22
AL381047 homologue to PIR|A86193|A86 hypothetical protein [impor... 34 0.22
TC85676 similar to GP|22655264|gb|AAM98222.1 unknown protein {Ar... 33 0.37
NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein 32 0.63
BI311119 similar to GP|8843772|db contains similarity to zinc fi... 32 0.63
AL385482 similar to GP|5106924|gb|A putative cell wall protein {... 32 0.82
BQ152925 weakly similar to GP|6448504|emb| Trihydrophobin {Clavi... 31 1.4
>TC86748 similar to GP|15485584|emb|CAC67503. SET-domain-containing protein
{Nicotiana tabacum}, partial (61%)
Length = 2742
Score = 926 bits (2393), Expect(2) = 0.0
Identities = 432/532 (81%), Positives = 485/532 (90%)
Frame = +1
Query: 165 DVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLMIYEVMRRKLGQIDEKAKGSHSG 224
DVD DAVA++IL++INP VF+++N PDGSRD+V YTLMIYEV+RRKLGQI+E K H+G
Sbjct: 742 DVDLDAVAHDILQSINPMVFDVINHPDGSRDSVTYTLMIYEVLRRKLGQIEESTKDLHTG 921
Query: 225 AKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLG 284
AKRPDLKAG +M TKG+R+NS+KRIG+VPGVE+GDIFFFRFE+CLVGLH+PSMAGIDYL
Sbjct: 922 AKRPDLKAGNVMMTKGVRSNSKKRIGIVPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLT 1101
Query: 285 TKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTSREKGASDQKLERGNLALERSL 344
+K SQEEEPLAVSIVSSGGYED+ DGDVLIYSGQGG +REKGASDQKLERGNLALE+S+
Sbjct: 1102 SKASQEEEPLAVSIVSSGGYEDDTGDGDVLIYSGQGGVNREKGASDQKLERGNLALEKSM 1281
Query: 345 HRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAY 404
HRGNDVRVIRG++D HP+GKVYVYDG+YKIQ+SWVEKAKSGFNVFKYKL R+ GQP+AY
Sbjct: 1282 HRGNDVRVIRGLKDVMHPSGKVYVYDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPEAY 1461
Query: 405 MIWKSILQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRL 464
IWKSI QWTDK+A R GVILPDLTSGAEK+PVCLVNDVDNEKGPAYFTY PTLKNL +
Sbjct: 1462 TIWKSIQQWTDKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTYIPTLKNLRGV 1641
Query: 465 APVESSEGCTCNGGCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSC 524
APVESS GC+C GGCQPG+ C C QKNGGYLPY+AAGL+ADLKSV++ECGPSC CPP+C
Sbjct: 1642 APVESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTC 1821
Query: 525 RNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDD 584
RNR+SQ GLK RLEVFRT KGWGLRSWD IRAGTFICEYAGEVIDNAR E L ENED+
Sbjct: 1822 RNRISQAGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDE 2001
Query: 585 YIFDSTRIYQQLEVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENK 644
YIFDSTRIYQQLEVF +++EAPKIPSPLYITA+NEGNVARFMNHSC+PNVLWRP+VRENK
Sbjct: 2002 YIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENK 2181
Query: 645 NEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLCGSVKCRGYFC 696
NE DLH+AF+AIRHIPPMMELTYDYGI LPL+ GQ+KK CLCGSVKCRGYFC
Sbjct: 2182 NEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAGQRKKNCLCGSVKCRGYFC 2337
Score = 221 bits (562), Expect(2) = 0.0
Identities = 112/149 (75%), Positives = 124/149 (83%), Gaps = 3/149 (2%)
Frame = +2
Query: 1 MDHNLGQDPAPAAGSFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGP 60
MDHNLGQ+ PA DKSRVLNVKPLRTLVPVFPSPSNPSSS+ PQGGAPFV VSP+GP
Sbjct: 221 MDHNLGQESVPA----DKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPAGP 388
Query: 61 FPSGVAPFYPFFVSPESQRLSEQNAQTPTAQRAAPISAAVPINSFRTPTGATNGDVGSSR 120
FP+GVAPFYPFFVSPESQRLSEQ+A PT QRA PISAAVPINSF+TPT ATNGDVGSSR
Sbjct: 389 FPAGVAPFYPFFVSPESQRLSEQHAPNPTPQRATPISAAVPINSFKTPTAATNGDVGSSR 568
Query: 121 RKS---RGQLPEDDNFVDLSEVDGEGGTG 146
RKS RGQL E++ + + +D + TG
Sbjct: 569 RKSRTRRGQLTEEEGYDNTEVIDVDAETG 655
>TC89006 weakly similar to GP|20466308|gb|AAM20471.1 unknown protein
{Arabidopsis thaliana}, partial (48%)
Length = 1715
Score = 534 bits (1375), Expect = e-152
Identities = 261/467 (55%), Positives = 334/467 (70%), Gaps = 21/467 (4%)
Frame = +3
Query: 250 GVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVE 309
G VPGVE+GDIFFFR E+C+VGLHA SM GID L + + EE LAVSIVSSG Y+D +
Sbjct: 3 GSVPGVEIGDIFFFRMEMCVVGLHAQSMGGIDALHIQGDRGEETLAVSIVSSGEYDDEAD 182
Query: 310 DGDVLIYSGQGGT--SREKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVY 367
DGDV+IY+GQGG ++K SDQKL +GNLAL+RS N++RVIRG++D +P K Y
Sbjct: 183 DGDVIIYTGQGGNFNKKDKHVSDQKLHKGNLALDRSSRTHNEIRVIRGIKDAVNPGAKTY 362
Query: 368 VYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPD 427
VYDGLYKIQ+SWVEKAK G +FKYKL+R+PGQP A+ +WKS+ +W ++ G+IL D
Sbjct: 363 VYDGLYKIQDSWVEKAKGGGGLFKYKLIRVPGQPSAFAVWKSVQKWKAGFPAKTGLILAD 542
Query: 428 LTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNG--GCQPGSHK 485
L+SGAE LPV LVN+VDN K PA+FTY +L++ + ++ S C+C+G C PG
Sbjct: 543 LSSGAESLPVSLVNEVDNVKSPAFFTYFHSLRHPKSFSLMQPSHSCSCSGKKACVPGDLD 722
Query: 486 CGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGK 545
C C ++N G PY G+LA+ K +V+ECGP+C C P+C+NRVSQ GLK ++EVF+TK K
Sbjct: 723 CSCIRRNEGDFPYIINGVLANRKPLVHECGPTCQCFPNCKNRVSQTGLKHQMEVFKTKDK 902
Query: 546 GWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENE-DDYIFDSTRIYQQLE------- 597
GWGLRSWDPIRAG FICEYAGEVID AR+ +L E + D+Y+FD+TRIY+ +
Sbjct: 903 GWGLRSWDPIRAGAFICEYAGEVIDKARLSQLVQEGDTDEYVFDTTRIYESFKWNYEPKL 1082
Query: 598 ----VFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAF 653
+ + E +P PL I A+N GNVARFMNHSC+PNV W+PV+ E N++ LHVAF
Sbjct: 1083LEEAITNESSEDYALPHPLIINAKNVGNVARFMNHSCSPNVFWQPVLYEENNQSFLHVAF 1262
Query: 654 YAIRHIPPMMELTYDYGI-----VLPLKVGQKKKKCLCGSVKCRGYF 695
+A+RHIPPM ELTYDYG + +KKCLCGS CRG F
Sbjct: 1263FALRHIPPMHELTYDYGSDRSDHTEGSSARKGRKKCLCGSSNCRGSF 1403
>TC92618 weakly similar to PIR|T02416|T02416 probable SET-domain
transcription regulator At2g23750 [imported] -
Arabidopsis thaliana, partial (77%)
Length = 781
Score = 115 bits (288), Expect = 6e-26
Identities = 71/187 (37%), Positives = 102/187 (53%), Gaps = 2/187 (1%)
Frame = +3
Query: 509 SVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEV 568
S+V+EC C C +C NR+ Q G++++LEVF T+ KG+G+R+ + I GTF+CEY GEV
Sbjct: 3 SLVFECNDKCGCNKTCPNRILQNGVRVKLEVFMTEKKGFGVRAGEAILRGTFVCEYIGEV 182
Query: 569 IDNARVEELSGENED-DYIFDSTRIYQQLEVFSSDVEAPKIPSPLY-ITARNEGNVARFM 626
++ G E+ Y D I + S VE P Y I + GNV+RF+
Sbjct: 183 LEQQEAHNRRGSKENCSYFLD---IDARANHTSRLVEG----HPRYVIDSTTYGNVSRFI 341
Query: 627 NHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLC 686
N+SC+PN++ V+ E + H+ YA R I ELT++Y G CLC
Sbjct: 342 NNSCSPNLVDYKVLVEATDCKHAHIGLYASRDIALGEELTFNYDYEPVPGEGD----CLC 509
Query: 687 GSVKCRG 693
GS+KC G
Sbjct: 510 GSLKC*G 530
>BE203534 similar to GP|10178033|dbj SET-domain protein-like {Arabidopsis
thaliana}, partial (12%)
Length = 294
Score = 108 bits (270), Expect = 7e-24
Identities = 54/95 (56%), Positives = 69/95 (71%), Gaps = 11/95 (11%)
Frame = +1
Query: 588 DSTRIYQQ---------LEVFSSDV--EAPKIPSPLYITARNEGNVARFMNHSCTPNVLW 636
D++RIY+ LE SS+V E IPSPL I+ARN GN+ARFMNHSC+PNV W
Sbjct: 1 DTSRIYEPFKWNYEPSLLEDVSSNVCSEDYTIPSPLIISARNVGNIARFMNHSCSPNVFW 180
Query: 637 RPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGI 671
+PV+ N++ +H+AF+A+RHIPPM ELTYDYGI
Sbjct: 181 QPVLYAENNQSFIHIAFFALRHIPPMAELTYDYGI 285
>TC93573 weakly similar to PIR|T02416|T02416 probable SET-domain
transcription regulator At2g23750 [imported] -
Arabidopsis thaliana, partial (58%)
Length = 908
Score = 84.0 bits (206), Expect = 2e-16
Identities = 55/152 (36%), Positives = 77/152 (50%), Gaps = 3/152 (1%)
Frame = +3
Query: 545 KGWGLRSWDPIRAGTFICEYAGEVIDNARVEELS---GENEDDYIFDSTRIYQQLEVFSS 601
KG G+R+ + I GTF+CEY GEV+D G Y +D I ++ S
Sbjct: 27 KGMGVRAGEAILRGTFVCEYIGEVLDVQEAHNRRKRYGTGNCSYFYD---INARVNDMSR 197
Query: 602 DVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPP 661
+E + I A GNV+RF+NHSC+PN++ V+ E+ + H+ FYA + I
Sbjct: 198 MIEEK---AQYVIDASKNGNVSRFINHSCSPNLVSHQVLVESMDCERSHIGFYASQDIAL 368
Query: 662 MMELTYDYGIVLPLKVGQKKKKCLCGSVKCRG 693
ELTY + L V + CLC S KCRG
Sbjct: 369 GEELTYGFQYEL---VPGEGSPCLCESSKCRG 455
>BG587693 weakly similar to GP|17066863|gb Su(VAR)3-9-related protein 4
{Arabidopsis thaliana}, partial (29%)
Length = 688
Score = 63.5 bits (153), Expect(2) = 7e-15
Identities = 46/147 (31%), Positives = 70/147 (47%), Gaps = 6/147 (4%)
Frame = +3
Query: 552 WDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSP 611
W + G F+CE+AGE++ + E + + ++ Y L D K
Sbjct: 126 WRNLPKGAFVCEFAGEILTIKELHERNIKCAEN----GKSTYPVLLDADWDSTFVKDEEA 293
Query: 612 LYITARNEGNVARFMNHSCTP-NVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYG 670
L + A + GN+ARF+NH C+ N++ P+ E + H A + R+I ELT+DYG
Sbjct: 294 LCLDAASFGNIARFINHRCSDANLVEIPIQIECPDRYYYHFALFTTRNIASHEELTWDYG 473
Query: 671 IVL-----PLKVGQKKKKCLCGSVKCR 692
I P+K+ Q C CGS CR
Sbjct: 474 IDFDDHDQPVKLFQ----CKCGSKFCR 542
Score = 35.4 bits (80), Expect(2) = 7e-15
Identities = 17/29 (58%), Positives = 21/29 (71%), Gaps = 1/29 (3%)
Frame = +2
Query: 524 CRNRVSQGGLKLRLEVFRT-KGKGWGLRS 551
C NRV Q G+ L+VF T +GKGWGLR+
Sbjct: 38 CGNRVIQRGITYNLQVFFTSEGKGWGLRT 124
>TC82595 similar to GP|10178033|dbj|BAB11516. SET-domain protein-like
{Arabidopsis thaliana}, partial (7%)
Length = 812
Score = 78.2 bits (191), Expect = 1e-14
Identities = 63/254 (24%), Positives = 109/254 (42%)
Frame = +3
Query: 1 MDHNLGQDPAPAAGSFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGP 60
M+ LGQ P GS DK ++L++KP+R+L+PVF S PQG SG
Sbjct: 210 MEEGLGQHSVPPPGSIDKYKILDIKPIRSLIPVF--------SKNPQG-------QSSGQ 344
Query: 61 FPSGVAPFYPFFVSPESQRLSEQNAQTPTAQRAAPISAAVPINSFRTPTGATNGDVGSSR 120
+PSG +PF+PF +S + T + + P+ +FR+P G
Sbjct: 345 YPSGFSPFFPFGGPHDS---------STTGAKPRRTAMPTPLQAFRSPFGEEE------- 476
Query: 121 RKSRGQLPEDDNFVDLSEVDGEGGTGDGKRRKPQKRIREKRCSSDVDPDAVANEILKTIN 180
DL++ D + + ++++ + +DV D L I+
Sbjct: 477 --------------DLNDNDDFSNKRSAASQSTRVKLKKHKVYNDVHVDLSG---LVGIS 605
Query: 181 PGVFEILNQPDGSRDAVAYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKG 240
PG + +G+R+ V LM ++ +RR+L Q+ + + + + K+ + +
Sbjct: 606 PG-----QRDNGNREVVNTVLMTFDALRRRLSQLVDAKELNTGFDQTYXFKSWQYLYDQR 770
Query: 241 IRANSRKRIGVVPG 254
KR+G VPG
Sbjct: 771 NSNKPTKRVGSVPG 812
>TC91822 similar to PIR|E96612|E96612 probable transcription factor
F12K22.14 [imported] - Arabidopsis thaliana, partial
(22%)
Length = 761
Score = 63.2 bits (152), Expect = 3e-10
Identities = 53/186 (28%), Positives = 83/186 (44%), Gaps = 10/186 (5%)
Frame = +1
Query: 295 AVSIVSSGGYEDNVEDGDVLIYSGQGGTSREKGASDQKLERGNLALERSLHRGNDVRVIR 354
A S+V SGGY + + G+ Y+G GG ++ D + N AL S +G VRV+R
Sbjct: 1 AQSVVLSGGYTQDEDHGEWFTYTGSGGRNQ---FLDHQFNNTNEALRLSCRKGYPVRVVR 171
Query: 355 GMRDEAH---PTGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSIL 411
+++ P V YDG+Y+I W E K+G V +Y VR +P
Sbjct: 172 SHKEKQSSYAPEAGVR-YDGVYRIDICWSEFGKNGEKVCRYLFVRCDNEP---------A 321
Query: 412 QWTDKSASRVGVILPDLTSGAEKLPVCLVN-----DVDNEKGPAYFTYSP--TLKNLNRL 464
WT + LP + + + + N D D EKG + P + + LN +
Sbjct: 322 PWTSDLSGDYPRTLPFIEEFRDAVDIIERNGDPSWDFDEEKGCWLWKKPPP*SKRPLNIV 501
Query: 465 APVESS 470
P+E++
Sbjct: 502 DPIENA 519
>BF647695 similar to GP|6006866|gb| hypothetical protein {Arabidopsis
thaliana}, partial (29%)
Length = 460
Score = 48.9 bits (115), Expect = 7e-06
Identities = 39/134 (29%), Positives = 57/134 (42%)
Frame = +3
Query: 560 FICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSPLYITARNE 619
F+ +YAGE++ + + D + R L V + + K L I A
Sbjct: 6 FLFQYAGELLTTTEAQRR--QQHYDELASRGRFSSALLVVREHLPSGKACLRLNIDATRI 179
Query: 620 GNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQ 679
GNVARF+NHSC L +VR + + F+A + I EL + YG + G
Sbjct: 180 GNVARFVNHSCDGGNLSTKLVR-STGALFPRLCFFASKDIQKDEELAFSYGEIRKRSNG- 353
Query: 680 KKKKCLCGSVKCRG 693
+ C C S C G
Sbjct: 354 --RLCHCNSPSCLG 389
>TC90124 similar to GP|17529304|gb|AAL38879.1 putative transcription factor
{Arabidopsis thaliana}, partial (34%)
Length = 1315
Score = 45.1 bits (105), Expect = 9e-05
Identities = 28/87 (32%), Positives = 45/87 (51%), Gaps = 2/87 (2%)
Frame = +2
Query: 317 SGQGGTSREKGASDQKLERGNLALERSLHRGNDVRVIRGMRDE--AHPTGKVYVYDGLYK 374
SG T++ + + DQ+ E N AL S +G VRV+R +++ A+ YDG+Y+
Sbjct: 20 SGNKRTNKNQ-SFDQQFENMNEALRLSCRKGYPVRVVRSHKEKRSAYAPEAGVRYDGVYR 196
Query: 375 IQNSWVEKAKSGFNVFKYKLVRLPGQP 401
I+ W + G V +Y VR +P
Sbjct: 197 IEKCWRKIGIQGHKVCRYLFVRCDNEP 277
>BI312377 similar to GP|8843772|dbj contains similarity to zinc finger
protein~gene_id:MYN8.4 {Arabidopsis thaliana}, partial
(7%)
Length = 583
Score = 45.1 bits (105), Expect = 9e-05
Identities = 25/81 (30%), Positives = 43/81 (52%)
Frame = +2
Query: 614 ITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVL 673
+ A ++GN+AR +NHSC PN R + + + + + A ++ ELTYDY +
Sbjct: 2 VDATDKGNIARLINHSCMPNCYARIM---SVGDDESRIVLIAKTNVSAGDELTYDY-LFD 169
Query: 674 PLKVGQKKKKCLCGSVKCRGY 694
P + + K C+C + CR +
Sbjct: 170 PDEPDEFKVPCMCKAPNCRKF 232
>CB892369 weakly similar to GP|18376303|em related to regulatory protein SET1
{Neurospora crassa}, partial (2%)
Length = 740
Score = 41.6 bits (96), Expect = 0.001
Identities = 29/97 (29%), Positives = 45/97 (45%)
Frame = +2
Query: 537 LEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL 596
L V+++ G GL + I G + EY GE++ + ++ + E +YI Y+
Sbjct: 458 LVVYKSGIHGLGLYTSQCIYRGRMVVEYVGEIVG----QRVADKREIEYISGRKLQYKSA 625
Query: 597 EVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPN 633
F I I A +G +ARF+NHSC PN
Sbjct: 626 CYFF*------IDKEHIIDATRKGGIARFVNHSCLPN 718
>BQ151164
Length = 772
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/43 (46%), Positives = 28/43 (64%)
Frame = +1
Query: 238 TKGIRANSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMAGI 280
TK +R++ RIG VP ++ +I FF LC+ G+HA SM GI
Sbjct: 256 TKRVRSDPPVRIGSVPLFQMENIVFFLIALCVGGMHALSMEGI 384
>BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania major}, partial
(1%)
Length = 766
Score = 33.9 bits (76), Expect = 0.22
Identities = 20/61 (32%), Positives = 29/61 (46%), Gaps = 5/61 (8%)
Frame = +1
Query: 35 PSPSNPSSSATPQGGAPFVCVSPS-----GPFPSGVAPFYPFFVSPESQRLSEQNAQTPT 89
P+P++P++ ATP G SPS GP S + P SP + S ++ TP
Sbjct: 172 PAPASPTAQATPPTGPSTAAPSPSTCTTTGPTSSSTSASSPAATSPRTTSPSPHSSGTPP 351
Query: 90 A 90
A
Sbjct: 352 A 354
>AL381047 homologue to PIR|A86193|A86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (5%)
Length = 490
Score = 33.9 bits (76), Expect = 0.22
Identities = 18/46 (39%), Positives = 22/46 (47%)
Frame = +3
Query: 648 DLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLCGSVKCRG 693
D H+ +A R I ELTYDY ++ C CG KCRG
Sbjct: 27 DEHIIIFAKRDIKQWEELTYDYRFFSI----DERLSCYCGFPKCRG 152
>TC85676 similar to GP|22655264|gb|AAM98222.1 unknown protein {Arabidopsis
thaliana}, partial (38%)
Length = 3105
Score = 33.1 bits (74), Expect = 0.37
Identities = 25/83 (30%), Positives = 31/83 (37%), Gaps = 5/83 (6%)
Frame = +2
Query: 10 APAAGSFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGG-----APFVCVSPSGPFPSG 64
APA + S + P PS S + +ATP G PF +P+ P SG
Sbjct: 2270 APAISASPSSISYPIIDFSGTAPAVPSFSGTAPAATPFSGTAPAATPFSGTAPAAPSSSG 2449
Query: 65 VAPFYPFFVSPESQRLSEQNAQT 87
AP P F S Q T
Sbjct: 2450 TAPAAPSFSGTAPAFHSNQQTST 2518
Score = 29.3 bits (64), Expect = 5.3
Identities = 25/73 (34%), Positives = 29/73 (39%), Gaps = 8/73 (10%)
Frame = +2
Query: 37 PSNPSSSATPQGGAPFVCVSPSG-PFP----SGVAPFYPFFVSPESQRLSEQNAQTP--- 88
P+N +S P AP + SPS +P SG AP P F A TP
Sbjct: 2231 PTNETSFVGPAASAPAISASPSSISYPIIDFSGTAPAVPSFSGTAP-------AATPFSG 2389
Query: 89 TAQRAAPISAAVP 101
TA A P S P
Sbjct: 2390 TAPAATPFSGTAP 2428
>NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein
Length = 576
Score = 32.3 bits (72), Expect = 0.63
Identities = 23/78 (29%), Positives = 31/78 (39%)
Frame = +1
Query: 36 SPSNPSSSATPQGGAPFVCVSPSGPFPSGVAPFYPFFVSPESQRLSEQNAQTPTAQRAAP 95
+P P++ G AP +P GP P G AP SP + T AP
Sbjct: 301 TPPAPAAPGAAPGAAPGTAPAPGGPPPEGAAP------SPAKGGAAAPTPGAGTGTSVAP 462
Query: 96 ISAAVPINSFRTPTGATN 113
A+ + +T TGA N
Sbjct: 463 AGAS-GSTAAKTATGAGN 513
>BI311119 similar to GP|8843772|db contains similarity to zinc finger
protein~gene_id:MYN8.4 {Arabidopsis thaliana}, partial
(10%)
Length = 798
Score = 32.3 bits (72), Expect = 0.63
Identities = 12/27 (44%), Positives = 18/27 (66%)
Frame = +2
Query: 607 KIPSPLYITARNEGNVARFMNHSCTPN 633
++ + I A + GN+AR +NHSC PN
Sbjct: 680 RLAREVVIDATDRGNIARLINHSCMPN 760
>AL385482 similar to GP|5106924|gb|A putative cell wall protein {Medicago
truncatula}, partial (42%)
Length = 402
Score = 32.0 bits (71), Expect = 0.82
Identities = 27/85 (31%), Positives = 36/85 (41%), Gaps = 3/85 (3%)
Frame = +2
Query: 33 VFPSPSN---PSSSATPQGGAPFVCVSPSGPFPSGVAPFYPFFVSPESQRLSEQNAQTPT 89
V P P+ P A P GGAP +P+GP P G AP +P ++ A TP
Sbjct: 56 VAPPPAGGAPPPGGAPPAGGAPPPGGAPAGPPPEGAAP------TP-----AKTAAPTPG 202
Query: 90 AQRAAPISAAVPINSFRTPTGATNG 114
+P++ A S P T G
Sbjct: 203 GATGSPVAPAGASGS-AAPKSPTTG 274
>BQ152925 weakly similar to GP|6448504|emb| Trihydrophobin {Claviceps
fusiformis}, partial (13%)
Length = 614
Score = 31.2 bits (69), Expect = 1.4
Identities = 25/81 (30%), Positives = 31/81 (37%)
Frame = +2
Query: 32 PVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGVAPFYPFFVSPESQRLSEQNAQTPTAQ 91
P P+ NP TP P V +P P PSG +P +PF P LS +
Sbjct: 272 PSTPTIPNPFQPPTP---TPLVPNNPFLPPPSGSSPLFPF---PSVPGLSPSXPPSSPPG 433
Query: 92 RAAPISAAVPINSFRTPTGAT 112
A P P TP +T
Sbjct: 434 LAFPFPPLFPPPGSGTPPAST 496
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,726,944
Number of Sequences: 36976
Number of extensions: 337262
Number of successful extensions: 2038
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 1973
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2018
length of query: 696
length of database: 9,014,727
effective HSP length: 103
effective length of query: 593
effective length of database: 5,206,199
effective search space: 3087276007
effective search space used: 3087276007
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0098b.1


