

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098a.6
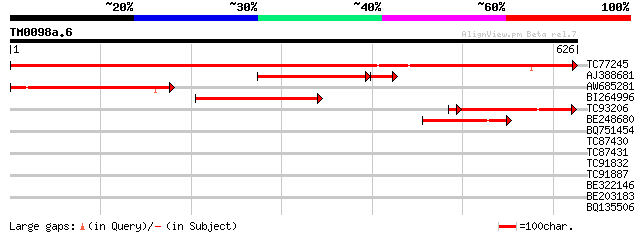
(626 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77245 similar to GP|6630556|gb|AAF19575.1| putative alpha-L-ar... 841 0.0
AJ388681 similar to GP|13398412|gb arabinoxylan arabinofuranohyd... 215 2e-63
AW685281 weakly similar to GP|17380938|gb putative arabinosidase... 235 3e-62
BI264996 weakly similar to GP|16417958|gb arabinosidase ARA-1 {L... 218 7e-57
TC93206 similar to GP|13398412|gb|AAK21879.1 arabinoxylan arabin... 146 1e-36
BE248680 weakly similar to GP|13937191|gb AT3g10740/T7M13_18 {Ar... 132 5e-31
BQ751454 weakly similar to GP|23326899|gb| alpha-L-arabinosidase... 40 0.002
TC87430 32 0.73
TC87431 32 0.73
TC91832 similar to GP|13925771|gb|AAK49438.1 phytase {Glycine ma... 30 2.1
TC91887 weakly similar to PIR|E96671|E96671 hypothetical protein... 29 6.2
BE322146 weakly similar to GP|20147226|gb| At2g44720/F16B22.21 {... 28 8.1
BE203183 28 8.1
BQ135506 28 8.1
>TC77245 similar to GP|6630556|gb|AAF19575.1| putative
alpha-L-arabinofuranosidase {Arabidopsis thaliana},
partial (89%)
Length = 2351
Score = 841 bits (2173), Expect = 0.0
Identities = 399/631 (63%), Positives = 496/631 (78%), Gaps = 5/631 (0%)
Frame = +2
Query: 1 CYVADAAGDQTSTLTVDLKSA-GRPIPETLFGVFYEEINHAGTGGLWAELVNNRGFEAGG 59
C+ D +TL VD A GR IPETLFG+F+EEINHAG GGLWAELV+NRGFEAGG
Sbjct: 275 CFDVQVQADLNATLVVDASQASGRRIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGG 454
Query: 60 TQTPSNIAPWTIVGKESLVLLQTELSSCFERNKVALRMDVLCD-QCPSDGVGVSNPGYWG 118
PSNI PW+I+G + + ++T+ +SCFERNKVALR++VLCD CP+DGVGV NPG+WG
Sbjct: 455 PNIPSNIDPWSIIGNATYINVETDRTSCFERNKVALRLEVLCDGTCPTDGVGVYNPGFWG 634
Query: 119 MNVVQKKEYKVVFFVKSTGSLDMTISFKKAEDGGILASQNVKASASEVANWKRMELKLVP 178
MN+ Q K+YKVVF+ +STG L++ +S + G LAS + SAS+ +NW ++E L
Sbjct: 635 MNIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSLASTVITGSASDFSNWTKVETVLEA 814
Query: 179 TASNPKATLHLTTTQKGVIWLDQVSAMPVDTFKGHGFRTDLVNMVLELKPAFIRFPGGCF 238
A+NP + L LTTT KGVIWLDQVSAMP+DT+KGHGFR+DL+ M+++LKP+FIRFPGGCF
Sbjct: 815 KATNPNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHGFRSDLLQMLVDLKPSFIRFPGGCF 994
Query: 239 IEGQTLRNAFRWKDSVGPWEERPGHFGDVWGSWTDEGLGYFEGLQLAEDIGAKPIWVFNN 298
+EG LRNAFRWK +VGPWEERPGHFGDVW WTD+GLGY+E LQL+ED+GA PIWVFNN
Sbjct: 995 VEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTDDGLGYYEFLQLSEDLGALPIWVFNN 1174
Query: 299 GISHTDQIDTSVITPFVQEALDGIEFARGPATSKWGSLRKSMGHPEPFDLKYVAIGNEDC 358
G+SH D++DTS + PFVQEALDG+EFARG TSKWGS+R +MGHPEPF+LKYVA+GNEDC
Sbjct: 1175GVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSKWGSMRAAMGHPEPFNLKYVAVGNEDC 1354
Query: 359 GKKNYLGNYLAFYNAIRKAYPDIQMISNCDASSKPLDHPADLYDYHTYPNNPSNMFNNAH 418
GKKNY GNYL FY+AIR+AYPDIQ+ISNCD SS+PLDHPAD+YDYH Y N ++MF+ +
Sbjct: 1355GKKNYRGNYLRFYDAIRRAYPDIQIISNCDGSSRPLDHPADMYDYHIY-TNANDMFSRST 1531
Query: 419 VFDKTPRKGPKAFVSEYALVGDQQAKLGTLIGGVSEAGFLIGLEKNSDHVAMAAYAPLFV 478
F++ R GPKAFVSEYA+ G+ A G+L+ ++EAGFLIGLEKNSD V MA+YAPLFV
Sbjct: 1532TFNRVTRSGPKAFVSEYAVTGN-DAGQGSLLAALAEAGFLIGLEKNSDIVHMASYAPLFV 1708
Query: 479 NADDRKWNPDAIVFNSNQVYGTPSYWVTHMFKESNGATFLASTLQTPDPSSFDASTILWQ 538
NA+DR+WNPDAIVFNS Q+YGTPSYW+ F ESNGAT L S+LQT +S AS I WQ
Sbjct: 1709NANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLNSSLQTTASNSLVASAITWQ 1888
Query: 539 NPQDKKTYLKIKVANLGNNQVKLGIVVHGLESSKII---GTKTVLTSKNALDENTFLEPR 595
N DKK Y++IK N G + V L I +GL+ + + TKTVLTS N +DEN+F +P+
Sbjct: 1889NSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQPK 2068
Query: 596 KIVPQQTPLEEASANMNVELPPLSVTSFDIL 626
K++P Q+ L+ +MNV +PP S TSFD+L
Sbjct: 2069KVIPIQSLLQSVGKDMNVIVPPHSFTSFDLL 2161
>AJ388681 similar to GP|13398412|gb arabinoxylan arabinofuranohydrolase
isoenzyme AXAH-I {Hordeum vulgare}, partial (18%)
Length = 574
Score = 215 bits (548), Expect(2) = 2e-63
Identities = 98/125 (78%), Positives = 114/125 (90%)
Frame = +1
Query: 274 EGLGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDTSVITPFVQEALDGIEFARGPATSKW 333
+GLG+FEGLQLAEDIGA PIWVFNNGISH+D++DTSVI+PFV+EAL+GIEFARG +TSKW
Sbjct: 1 DGLGFFEGLQLAEDIGALPIWVFNNGISHSDEVDTSVISPFVKEALEGIEFARGSSTSKW 180
Query: 334 GSLRKSMGHPEPFDLKYVAIGNEDCGKKNYLGNYLAFYNAIRKAYPDIQMISNCDASSKP 393
GS+R SMGHP+PF+LKYVAIGNEDC KKNY GNY+AFY AI+K YPDIQ+ISNC A P
Sbjct: 181 GSVRASMGHPKPFNLKYVAIGNEDCYKKNYYGNYMAFYKAIKKFYPDIQIISNCPAFKTP 360
Query: 394 LDHPA 398
L+HPA
Sbjct: 361 LNHPA 375
Score = 45.8 bits (107), Expect(2) = 2e-63
Identities = 19/30 (63%), Positives = 21/30 (69%)
Frame = +2
Query: 399 DLYDYHTYPNNPSNMFNNAHVFDKTPRKGP 428
DLYDYHTYP + MFN H FDK+P GP
Sbjct: 377 DLYDYHTYPIDARXMFNAYHEFDKSPXHGP 466
>AW685281 weakly similar to GP|17380938|gb putative arabinosidase
{Arabidopsis thaliana}, partial (18%)
Length = 651
Score = 235 bits (600), Expect = 3e-62
Identities = 117/184 (63%), Positives = 143/184 (77%), Gaps = 3/184 (1%)
Frame = +2
Query: 2 YVADAAGDQTSTLTVDLKSAGRPIPETLFGVFYEEINHAGTGGLWAELVNNRGFEAGGTQ 61
+ +A G Q S+L V+ + GRP+P TLFG+FYEEINHAG+GG+WA+LVNN GFEA GT+
Sbjct: 89 FQCNANGSQISSLVVNA-AQGRPMPNTLFGIFYEEINHAGSGGIWAQLVNNSGFEAAGTR 265
Query: 62 TPSNIAPWTIVGKESLVLLQTELSSCFERNKVALRMDVLCDQCPSDGVGVSNPGYWGMNV 121
TPSNI PWTI+G ES V LQTELSSCFERNKVALRMDVLCD+CP DGVGVSNPG+WGMN+
Sbjct: 266 TPSNIFPWTIIGTESSVKLQTELSSCFERNKVALRMDVLCDKCPPDGVGVSNPGFWGMNI 445
Query: 122 VQKKEYKVVFFVKSTGSLDMTISFKKAEDGGILASQNV---KASASEVANWKRMELKLVP 178
VQ K+YKVVFF +S GSLDM ++F+ A G ILAS ++ KAS V + ++ L
Sbjct: 446 VQGKKYKVVFFYRSLGSLDMRVAFRDAIXGRILASSHIIRHKASKKRVPKXQXVQTILEA 625
Query: 179 TASN 182
AS+
Sbjct: 626 RASS 637
>BI264996 weakly similar to GP|16417958|gb arabinosidase ARA-1 {Lycopersicon
esculentum}, partial (20%)
Length = 422
Score = 218 bits (554), Expect = 7e-57
Identities = 92/140 (65%), Positives = 113/140 (80%)
Frame = +2
Query: 206 PVDTFKGHGFRTDLVNMVLELKPAFIRFPGGCFIEGQTLRNAFRWKDSVGPWEERPGHFG 265
P+DT+KGHGFR +L MV ELKP RFPGGC++EG L+NAF+WK ++GPWE RPGH+G
Sbjct: 2 PLDTYKGHGFRMNLFQMVAELKPRXFRFPGGCYVEGNVLKNAFQWKQTIGPWENRPGHYG 181
Query: 266 DVWGSWTDEGLGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDTSVITPFVQEALDGIEFA 325
DVW WTD+G G+FEGLQLAED+ A PIWVFNNGISH++Q++ S I+P VQ ALDGIE A
Sbjct: 182 DVWDYWTDDGFGFFEGLQLAEDLNALPIWVFNNGISHSEQVNVSAISPSVQXALDGIEXA 361
Query: 326 RGPATSKWGSLRKSMGHPEP 345
G TS+WGS+ MGHP+P
Sbjct: 362 IGSPTSRWGSIIAXMGHPKP 421
>TC93206 similar to GP|13398412|gb|AAK21879.1 arabinoxylan
arabinofuranohydrolase isoenzyme AXAH-I {Hordeum
vulgare}, partial (6%)
Length = 642
Score = 146 bits (368), Expect(2) = 1e-36
Identities = 76/132 (57%), Positives = 94/132 (70%), Gaps = 1/132 (0%)
Frame = +1
Query: 495 NQVYGTPSYWVTHMFKESNGATFLASTLQTPDPSSFDASTILWQNPQDKKTYLKIKVANL 554
+QVYGTPSYWVT++FKESNGATFL STLQT DP + AS IL ++PQ+K TYLKIK+AN+
Sbjct: 37 HQVYGTPSYWVTYLFKESNGATFLNSTLQTTDPGTLAASAILVKDPQNKNTYLKIKIANM 216
Query: 555 GNNQVKLGIVVHGLESSKIIG-TKTVLTSKNALDENTFLEPRKIVPQQTPLEEASANMNV 613
QV I + G S + G TKTVLT N LDEN+F EP+KI PQ +PL+ MNV
Sbjct: 217 RKTQVDFKISIQGFASKNLKGSTKTVLTG-NELDENSFAEPKKIAPQTSPLQNPGNEMNV 393
Query: 614 ELPPLSVTSFDI 625
+ P S+T D+
Sbjct: 394 IVQPTSLTILDM 429
Score = 25.8 bits (55), Expect(2) = 1e-36
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = +2
Query: 485 WNPDAIVFNSNQVYG 499
WNPDAIVF+S + G
Sbjct: 8 WNPDAIVFSSIKFMG 52
>BE248680 weakly similar to GP|13937191|gb AT3g10740/T7M13_18 {Arabidopsis
thaliana}, partial (15%)
Length = 375
Score = 132 bits (331), Expect = 5e-31
Identities = 66/99 (66%), Positives = 78/99 (78%)
Frame = +1
Query: 456 GFLIGLEKNSDHVAMAAYAPLFVNADDRKWNPDAIVFNSNQVYGTPSYWVTHMFKESNGA 515
GFLIGLEKNSD V+M YAPLFVN +DRKW PDAIVF+S+QVYG PSYW+ +FKES+GA
Sbjct: 76 GFLIGLEKNSDVVSMVNYAPLFVNTNDRKWTPDAIVFDSHQVYGIPSYWLIKLFKESSGA 255
Query: 516 TFLASTLQTPDPSSFDASTILWQNPQDKKTYLKIKVANL 554
TFL STLQT P + AS I W++ D + L+IKVANL
Sbjct: 256 TFLNSTLQTDSP-TLAASAISWKSSVDGTSILRIKVANL 369
>BQ751454 weakly similar to GP|23326899|gb| alpha-L-arabinosidase
{Bifidobacterium longum NCC2705}, partial (12%)
Length = 780
Score = 40.4 bits (93), Expect = 0.002
Identities = 21/48 (43%), Positives = 26/48 (53%)
Frame = +3
Query: 214 GFRTDLVNMVLELKPAFIRFPGGCFIEGQTLRNAFRWKDSVGPWEERP 261
GFR D+V V EL +R+PGG F + W D VGP E+RP
Sbjct: 255 GFRKDVVEAVKELNCPVMRYPGGNFCA------TYHWIDGVGPREKRP 380
>TC87430
Length = 649
Score = 32.0 bits (71), Expect = 0.73
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = +3
Query: 235 GGCFIEGQTLRNAFRWKDSVG 255
GGCF+E LRN +WK +VG
Sbjct: 459 GGCFVERNYLRNEIQWKAAVG 521
>TC87431
Length = 662
Score = 32.0 bits (71), Expect = 0.73
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = +2
Query: 235 GGCFIEGQTLRNAFRWKDSVG 255
GGCF+E LRN +WK +VG
Sbjct: 437 GGCFVERNYLRNEIQWKAAVG 499
>TC91832 similar to GP|13925771|gb|AAK49438.1 phytase {Glycine max}, partial
(49%)
Length = 1204
Score = 30.4 bits (67), Expect = 2.1
Identities = 17/44 (38%), Positives = 23/44 (51%)
Frame = +3
Query: 397 PADLYDYHTYPNNPSNMFNNAHVFDKTPRKGPKAFVSEYALVGD 440
P LY Y + S M ++ H F P GPK++ S A+VGD
Sbjct: 756 PNTLYQYQCGDPSLSAM-SDVHYFRTMPVSGPKSYPSRIAVVGD 884
>TC91887 weakly similar to PIR|E96671|E96671 hypothetical protein F13O11.13
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1011
Score = 28.9 bits (63), Expect = 6.2
Identities = 19/49 (38%), Positives = 31/49 (62%), Gaps = 1/49 (2%)
Frame = +3
Query: 546 YLKIKVANLGNNQVKLGIVVHG-LESSKIIGTKTVLTSKNALDENTFLE 593
YL ++ ++GN +V++G V +G E + II + T LTS D+ +FLE
Sbjct: 498 YLTLEAFSVGNRRVEIGGVPNGDNEGNIIIDSGTTLTSLTK-DDYSFLE 641
>BE322146 weakly similar to GP|20147226|gb| At2g44720/F16B22.21 {Arabidopsis
thaliana}, partial (3%)
Length = 524
Score = 28.5 bits (62), Expect = 8.1
Identities = 13/49 (26%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Frame = +1
Query: 569 ESSKIIGTKTVLTSKNALDENTFLEPRKIVPQQTP-LEEASANMNVELP 616
+ S+++ KTV+ + ++EN +E + +V + P +E+ + +MN P
Sbjct: 229 QQSELVEEKTVIEEEIKVEENLVVEEKPVVEEDNPVVEDKAIDMNQIAP 375
>BE203183
Length = 408
Score = 28.5 bits (62), Expect = 8.1
Identities = 10/17 (58%), Positives = 12/17 (69%)
Frame = +2
Query: 235 GGCFIEGQTLRNAFRWK 251
GGCF+E LRN +WK
Sbjct: 356 GGCFVERNYLRNEIQWK 406
>BQ135506
Length = 890
Score = 28.5 bits (62), Expect = 8.1
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Frame = +3
Query: 544 KTYLKIKVANLGNNQVKLGIVV-HGLESSKIIGTKTVLTSKN 584
+T KIK N+G+ K+GI++ HG S I TKT+ + N
Sbjct: 207 RTLKKIKPVNMGDEIEKMGILMFHGWVLSTSI*TKTMRDANN 332
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,061,642
Number of Sequences: 36976
Number of extensions: 267491
Number of successful extensions: 1158
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1144
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1151
length of query: 626
length of database: 9,014,727
effective HSP length: 102
effective length of query: 524
effective length of database: 5,243,175
effective search space: 2747423700
effective search space used: 2747423700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0098a.6


