

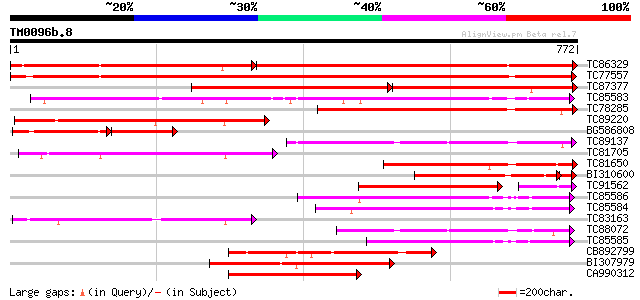
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0096b.8
(772 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase ... 664 0.0
TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 977 0.0
TC87377 weakly similar to PIR|T07171|T07171 subtilisin-like prot... 466 0.0
TC85583 similar to PIR|T51335|T51335 subtilisin-like proteinase ... 518 e-147
TC78285 similar to PIR|A84473|A84473 probable serine proteinase ... 355 3e-98
TC89220 similar to PIR|T05768|T05768 subtilisin-like proteinase ... 320 1e-87
BG586808 weakly similar to PIR|T07171|T07 subtilisin-like protei... 172 9e-87
TC89137 similar to GP|4200338|emb|CAA76726.1 P69C protein {Lycop... 280 2e-75
TC81705 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428... 264 1e-70
TC81650 similar to PIR|T07171|T07171 subtilisin-like proteinase ... 251 1e-66
BI310600 similar to PIR|S52770|S52 subtilisin-like proteinase (E... 241 2e-66
TC91562 similar to PIR|T07172|T07172 subtilisin-like proteinase ... 215 7e-65
TC85586 similar to GP|20521377|dbj|BAB91889. putative subtilisin... 242 4e-64
TC85584 weakly similar to PIR|A84473|A84473 probable serine prot... 231 6e-61
TC83163 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428... 229 3e-60
TC88072 similar to PIR|T06580|T06580 subtilisin-like proteinase ... 228 9e-60
TC85585 similar to GP|22773236|gb|AAN06842.1 Putatvie subtilisin... 224 7e-59
CB892799 weakly similar to PIR|T05768|T05 subtilisin-like protei... 216 3e-56
BI307979 weakly similar to GP|20521377|db putative subtilisin-li... 214 1e-55
CA990312 weakly similar to GP|14488360|gb putative serine protea... 214 1e-55
>TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase (EC 3.4.21.-)
1 - tomato, partial (91%)
Length = 2671
Score = 664 bits (1714), Expect(2) = 0.0
Identities = 332/437 (75%), Positives = 365/437 (82%)
Frame = +3
Query: 336 TTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYAGNASNFSVGYLCLPD 395
TTVGA TIDRDFPAYITLGN + G SLY GK +SPLPLVYA N S S LC D
Sbjct: 1083 TTVGARTIDRDFPAYITLGNGNRYNGVSLYNGKLPPNSPLPLVYAANVSQDSSDNLCSTD 1262
Query: 396 SLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAAL 455
SL+PSKV GKIVIC+RGGN R EK LVVKRAGGIGMILANN+++GEELVADS LLPAAAL
Sbjct: 1263 SLIPSKVSGKIVICDRGGNPRAEKSLVVKRAGGIGMILANNQDYGEELVADSFLLPAAAL 1442
Query: 456 GERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAP 515
GE++S +K Y S+ NPTAK+ FGGT V+PSPVVAAFSSRGPN LTPKILKPDLIAP
Sbjct: 1443 GEKASNEIKKYASSAPNPTAKIAFGGTRFGVQPSPVVAAFSSRGPNILTPKILKPDLIAP 1622
Query: 516 GVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRS 575
GVNILAGW+G +GPTGL VDTRHVSFNIISGTSMSCPHVSGLAA+LKG+HPEWSPAAIRS
Sbjct: 1623 GVNILAGWSGKVGPTGLSVDTRHVSFNIISGTSMSCPHVSGLAALLKGAHPEWSPAAIRS 1802
Query: 576 ALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCA 635
ALMTTSY YKNGQTI+DVATG PATPLD+G+GHVDPVA+LDPGLVYDA DDYL FLCA
Sbjct: 1803 ALMTTSYGTYKNGQTIKDVATGIPATPLDYGSGHVDPVAALDPGLVYDATTDDYLNFLCA 1982
Query: 636 LNYTSLEIKLASRRDFKCDPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTL 695
LNY S +IKL +RR+F CD + KYRVED NYPSF+VP +TASG G SH P V+Y R L
Sbjct: 1983 LNYNSFQIKLVARREFTCDKRIKYRVEDLNYPSFSVPFDTASG-RGSSHNPSIVQYKRIL 2159
Query: 696 TNVGTPGTYKASVSSQSPSVKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKSFAYLY 755
TNVG P TYK SVSSQSP KI VEPQ L F+EL EKKSYTVTFTS+SMPSGT SFA+L
Sbjct: 2160 TNVGAPSTYKVSVSSQSPLDKIVVEPQTLSFKELNEKKSYTVTFTSHSMPSGTTSFAHLE 2339
Query: 756 WSDGKHRVASPIAITWT 772
WSDGKH+V SPIA +WT
Sbjct: 2340 WSDGKHKVTSPIAFSWT 2390
Score = 482 bits (1241), Expect(2) = 0.0
Identities = 238/338 (70%), Positives = 278/338 (81%), Gaps = 3/338 (0%)
Frame = +2
Query: 1 MKMPIFQLLQSALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFDS 60
M M IF+ LQ ALLL+L S +TIAEKKT Q K+TYIIHMDK MPA+F+DH W+DS
Sbjct: 80 MNMLIFKCLQMALLLVL--SSRFTIAEKKT-QHLKRTYIIHMDKFNMPASFDDHLQWYDS 250
Query: 61 SLQSVSESAEILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFL 120
SL+SVS++AE +YTYKHVAHGFSTRLT QEA+ L +QPG+LSV P+VRYELHTTRTPEFL
Sbjct: 251 SLKSVSDTAETMYTYKHVAHGFSTRLTTQEADLLTKQPGILSVIPDVRYELHTTRTPEFL 430
Query: 121 GLLKKTTTLSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSS 180
GL +KT TL P S KQS+V++GV+DTGVWPELKS DDTGL PVP +WKG+CE G NSS
Sbjct: 431 GL-EKTITLLPSSGKQSEVIVGVIDTGVWPELKSFDDTGLGPVPKSWKGECETGKTFNSS 607
Query: 181 SCNRKLIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLA 240
+CN+KL+GARFF+KGYEA GPID +TES+S RDDDGHGSHT TTAAGSAVAGASLFG A
Sbjct: 608 NCNKKLVGARFFAKGYEAAFGPIDENTESKSPRDDDGHGSHTSTTAAGSAVAGASLFGFA 787
Query: 241 SGTARGMATQARVAAYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSI---GGSSADYFR 297
SGTA+GMATQARVAAYKVCWLGGCF+SDIAA IDKAI+ + + GG DY++
Sbjct: 788 SGTAKGMATQARVAAYKVCWLGGCFTSDIAAAIDKAIDTQARTAAYKVSRLGGGLTDYYK 967
Query: 298 DIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWI 335
D +A+G F A HGILVS+SAGNGGPS +SL+N APWI
Sbjct: 968 DTVAMGTFAAIEHGILVSSSAGNGGPSKASLANVAPWI 1081
>TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (88%)
Length = 2682
Score = 977 bits (2526), Expect = 0.0
Identities = 486/773 (62%), Positives = 590/773 (75%), Gaps = 2/773 (0%)
Frame = +3
Query: 1 MKMPIFQLLQSALLLLLI-FCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFD 59
MK + +A+L+L + C + + K TYI+HM KS MP +F H W++
Sbjct: 240 MKFSFHRSFPTAILVLFMGLCDA--------SSSLKSTYIVHMAKSEMPESFEHHTLWYE 395
Query: 60 SSLQSVSESAEILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEF 119
SSLQSVS+SAE++YTY++ HGFSTRLT +EA L Q G+L+V PEV+YELHTTRTP+F
Sbjct: 396 SSLQSVSDSAEMMYTYENAIHGFSTRLTPEEARLLESQTGILAVLPEVKYELHTTRTPQF 575
Query: 120 LGLLKKTTTLSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNS 179
LGL K+ + P S ++VV+GVLDTGVWPE KS +D G P+P+TWKG CE+G N +
Sbjct: 576 LGL-DKSADMFPESSSGNEVVVGVLDTGVWPESKSFNDAGFGPIPTTWKGACESGTNFTA 752
Query: 180 SSCNRKLIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGL 239
++CN+KLIGARFFSKG EA LGPID +TES+S RDDDGHG+HT +TAAGS V ASLFG
Sbjct: 753 ANCNKKLIGARFFSKGVEAMLGPIDETTESKSPRDDDGHGTHTSSTAAGSVVPDASLFGY 932
Query: 240 ASGTARGMATQARVAAYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGGSSADYFRDI 299
ASGTARGMAT+ARVA YKVCW GGCFSSDI A IDKAI D VN++S+S+GG +DYFRD
Sbjct: 933 ASGTARGMATRARVAVYKVCWKGGCFSSDILAAIDKAISDNVNVLSLSLGGGMSDYFRDS 1112
Query: 300 IAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITH 359
+AIGAF+A GILVS SAGN GPS SLSN APWITTVGAGT+DRDFPA ++LGN + +
Sbjct: 1113VAIGAFSAMEKGILVSCSAGNAGPSAYSLSNVAPWITTVGAGTLDRDFPASVSLGNGLNY 1292
Query: 360 TGASLYRGKPLSDSPLPLVYAGNASNFSVGYLCLPDSLVPSKVLGKIVICERGGNARVEK 419
+G SLYRG L +SPLPL+YAGNA+N + G LC+ +L P V GKIV+C+RG NARV+K
Sbjct: 1293SGVSLYRGNALPESPLPLIYAGNATNATNGNLCMTGTLSPELVAGKIVLCDRGMNARVQK 1472
Query: 420 GLVVKRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVF 479
G VVK AGG+GM+L+N GEELVAD+HLLPA A+GER A+K Y+FS PT K+VF
Sbjct: 1473GAVVKAAGGLGMVLSNTAANGEELVADTHLLPATAVGEREGNAIKKYLFSEAKPTVKIVF 1652
Query: 480 GGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHV 539
GT + V+PSPVVAAFSSRGPN +TP+ILKPDLIAPGVNILAGW+ A+GPTGL VD R V
Sbjct: 1653QGTKVGVEPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAVGPTGLAVDERRV 1832
Query: 540 SFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKP 599
FNIISGTSMSCPHVSGLAA++K +HP+WSPAA+RSALMTT+Y AYKNG +QD ATGK
Sbjct: 1833DFNIISGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTTAYIAYKNGNKLQDSATGKS 2012
Query: 600 ATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCDPKKKY 659
+TP D G+GHVDPVA+L+PGLVYD DDYLGFLCALNYT+ +I +RR F+CD KKY
Sbjct: 2013STPFDHGSGHVDPVAALNPGLVYDLTADDYLGFLCALNYTATQITSLARRKFQCDAGKKY 2192
Query: 660 RVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKASVSSQSPSVKIAV 719
V D NYPSFAV +T G VK++R TNVG GTYKASV+S S +VKI V
Sbjct: 2193SVSDLNYPSFAVVFDTMGGAN-------VVKHTRIFTNVGPAGTYKASVTSDSKNVKITV 2351
Query: 720 EPQILRFQELYEKKSYTVTFTSN-SMPSGTKSFAYLYWSDGKHRVASPIAITW 771
EP+ L F + EKKS+TVTFTS+ S P F L W++GK+ V SPI+I+W
Sbjct: 2352EPEELSF-KANEKKSFTVTFTSSGSTPQKLNGFGRLEWTNGKNVVGSPISISW 2507
>TC87377 weakly similar to PIR|T07171|T07171 subtilisin-like proteinase (EC
3.4.21.-) 1 - tomato, partial (54%)
Length = 2071
Score = 466 bits (1198), Expect(2) = 0.0
Identities = 230/274 (83%), Positives = 250/274 (90%)
Frame = +2
Query: 248 ATQARVAAYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTA 307
AT+ARVAAYKVCWL GCF+SDIAAG+DKAIEDGVNI+SMSIGGS DY+RDIIAIGAFTA
Sbjct: 2 ATEARVAAYKVCWLSGCFTSDIAAGMDKAIEDGVNILSMSIGGSIMDYYRDIIAIGAFTA 181
Query: 308 NSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRG 367
SHGILVS+SAGNGGPS SLSN APWITTVGAGTIDRDFP+YITLGN T+TGASLY G
Sbjct: 182 MSHGILVSSSAGNGGPSAESLSNVAPWITTVGAGTIDRDFPSYITLGNGKTYTGASLYNG 361
Query: 368 KPLSDSPLPLVYAGNASNFSVGYLCLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAG 427
KP SDS LP+VYAGN S SVGYLC+PDSL SKVLGKIVICERGGN+RVEKGLVVK AG
Sbjct: 362 KPSSDSLLPVVYAGNVSESSVGYLCIPDSLTSSKVLGKIVICERGGNSRVEKGLVVKNAG 541
Query: 428 GIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVK 487
G+GMIL NNE +GEEL+ADSHLLPAAALG++SS LKDYVF+++NP AKLVFGGTHLQV+
Sbjct: 542 GVGMILVNNEAYGEELIADSHLLPAAALGQKSSTVLKDYVFTTKNPRAKLVFGGTHLQVQ 721
Query: 488 PSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILA 521
PSPVVAAFSSRGPN LTPKILKPDLIAPGVNILA
Sbjct: 722 PSPVVAAFSSRGPNSLTPKILKPDLIAPGVNILA 823
Score = 406 bits (1043), Expect(2) = 0.0
Identities = 192/254 (75%), Positives = 219/254 (85%), Gaps = 3/254 (1%)
Frame = +1
Query: 522 GWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTS 581
GWTGA+GPTGL +D RHV+FNIISGTSMSCPH SGLAAI+KG++PEWSPAAIRSALMTT+
Sbjct: 826 GWTGAVGPTGLALDKRHVNFNIISGTSMSCPHASGLAAIVKGAYPEWSPAAIRSALMTTA 1005
Query: 582 YTAYKNGQTIQDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSL 641
YT+YKNGQTI DVATGKPATP DFG+GHVDPV++LDPGLVYD NVDDYLGF CALNYTS
Sbjct: 1006 YTSYKNGQTIVDVATGKPATPFDFGSGHVDPVSALDPGLVYDINVDDYLGFFCALNYTSY 1185
Query: 642 EIKLASRRDFKCDPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTP 701
+IKLA+RR+F CD +KKYRVEDFNYPSFAV LETASGIGGGS+ PI V+Y+R LTNVG P
Sbjct: 1186 QIKLAARREFTCDARKKYRVEDFNYPSFAVALETASGIGGGSNKPIIVEYNRVLTNVGAP 1365
Query: 702 GTYKASV---SSQSPSVKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKSFAYLYWSD 758
GTY A+V S S SVK+ VEP+ + F+E+YEKK Y V F SMPSGTKSF YL W+D
Sbjct: 1366 GTYNATVVLSSVDSSSVKVVVEPETISFKEVYEKKGYKVRFICGSMPSGTKSFGYLEWND 1545
Query: 759 GKHRVASPIAITWT 772
GKH+V SPIA +WT
Sbjct: 1546 GKHKVGSPIAFSWT 1587
>TC85583 similar to PIR|T51335|T51335 subtilisin-like proteinase AIR3
auxin-induced [imported] - Arabidopsis thaliana
(fragment), partial (40%)
Length = 2691
Score = 518 bits (1335), Expect = e-147
Identities = 328/795 (41%), Positives = 447/795 (55%), Gaps = 54/795 (6%)
Frame = +2
Query: 29 KTAQQAKKTYIIHMDKST---------MPATFNDHQHWFDSSLQSVSESAE-ILYTYKHV 78
+T KK YI+++ + + N H S+L S ++ E I+Y+Y
Sbjct: 95 ETVHGTKKCYIVYLGAHSHGPRPTSLELEIATNSHYDLLSSTLGSREKAKEAIIYSYNKH 274
Query: 79 AHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLK--KTTTLSPGSDKQ 136
+GF+ L +EA +A++ V+SV ++LHTTR+ EFLGL + K T G +
Sbjct: 275 INGFAALLEDEEAADIAKKRNVVSVFLSKPHKLHTTRSWEFLGLRRNAKNTAWQKGKFGE 454
Query: 137 SQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKG--QCEAG--NNMNSSSCNRKLIGARFF 192
+ ++ + DTGVWPE KS +D G PVPS W+G CE + + CNRKLIGARFF
Sbjct: 455 NTIIANI-DTGVWPESKSFNDKGYGPVPSKWRGGKACEISKFSKYKKNPCNRKLIGARFF 631
Query: 193 SKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQAR 252
S YEA + + R+ARD GHG+HTL+TA G+ V AS+F + +GT +G + +AR
Sbjct: 632 SNAYEAYNDKLP--SWQRTARDFLGHGTHTLSTAGGNFVPDASVFAIGNGTVKGGSPRAR 805
Query: 253 VAAYKVCW----LGGCFSSDIAAGIDKAIEDGVNIISMSIGGSSA----DYFRDIIAIGA 304
VA YKVCW L CF +D+ A ID+AI DGV+IIS+S+ G S D F D ++IGA
Sbjct: 806 VATYKVCWSLLDLEDCFGADVLAAIDQAISDGVDIISLSLAGHSLVYPEDIFTDEVSIGA 985
Query: 305 FTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASL 364
F A S IL+ SAGN GP+ S+ N APW+ T+ A T+DRDF + IT+GN T GASL
Sbjct: 986 FHALSRNILLVASAGNEGPTGGSVVNVAPWVFTIAASTLDRDFSSTITIGNQ-TIRGASL 1162
Query: 365 YRGKPLSDSPLPLVYA-----GNASNFSVGYLCLPDSLVPSKVLGKIVICERGGNAR-VE 418
+ P + + PL+ + NA+N + C P +L PSKV GKIV C R GN + V
Sbjct: 1163FVNLPPNQA-FPLIVSTDGKLANATNHDAQF-CKPGTLDPSKVKGKIVECIREGNIKSVA 1336
Query: 419 KGLVVKRAGGIGMILANNEEFGEELVADSHLLPA---------------AALGERSSKAL 463
+G AG GM+L+N + G+ +A+ H L +A ER+
Sbjct: 1337EGQEALSAGAKGMLLSNQPKQGKTTLAEPHTLSCVEVPHHAPKPPKPKKSAEQERAGSHA 1516
Query: 464 KDYVFSSRNPTAK----LVFGG--THLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGV 517
+ +S + K + F G T KP+PV+A+FSSRGPN + P ILKPD+ APGV
Sbjct: 1517PAFDITSMDSKLKAGTTIKFSGAKTLYGRKPAPVMASFSSRGPNKIQPSILKPDVTAPGV 1696
Query: 518 NILAGWTGAIGPTGLPVDTRH-VSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSA 576
NILA ++ + L D R+ FN++ GTSMSCPHV+G+A ++K HP WSPAAI+SA
Sbjct: 1697NILAAYSLYASASNLKTDNRNNFPFNVLQGTSMSCPHVAGIAGLIKTLHPNWSPAAIKSA 1876
Query: 577 LMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCAL 636
+MTT+ T + IQD K A P D+G+GHV P ++DPGLVYD + DYL FLCA
Sbjct: 1877IMTTATTLDNTNRPIQDAFENKLAIPFDYGSGHVQPDLAIDPGLVYDLGIKDYLNFLCAY 2056
Query: 637 NYTSLEIK-LASRRDFKCDPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTL 695
Y I L F C + + DFNYPS +P + V +RT+
Sbjct: 2057GYNQQLISALNFNGTFIC--SGSHSITDFNYPSITLPNLKLN----------AVNVTRTV 2200
Query: 696 TNVGTPGTYKASVSSQSPSVKIAVEPQILRFQELYEKKSYTVTF-TSNSMPSGTKSFAYL 754
TNVG PGTY S +Q KI V P L F++ EKK++ V +N P G F L
Sbjct: 2201TNVGPPGTY--SAKAQLLGYKIVVLPNSLTFKKTGEKKTFQVIVQATNVTPRGKYQFGNL 2374
Query: 755 YWSDGKHRVASPIAI 769
W+DGKH V SPI +
Sbjct: 2375QWTDGKHIVRSPITV 2419
>TC78285 similar to PIR|A84473|A84473 probable serine proteinase [imported]
- Arabidopsis thaliana, partial (41%)
Length = 1353
Score = 355 bits (912), Expect = 3e-98
Identities = 190/357 (53%), Positives = 235/357 (65%), Gaps = 3/357 (0%)
Frame = +1
Query: 419 KGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLV 478
KG VVK AGGIGMILAN GEELVADSHLLPA A+G ++ YV S NPT L
Sbjct: 13 KGRVVKEAGGIGMILANTAASGEELVADSHLLPAVAVGRIIGDQIRKYVSSDLNPTTVLS 192
Query: 479 FGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRH 538
FGGT L V+PSPVVAAFSSRGPN +T +ILKPD+I PGVNILAGW+ A+GP+GL DTR
Sbjct: 193 FGGTVLNVRPSPVVAAFSSRGPNMITKEILKPDVIGPGVNILAGWSEAVGPSGLAEDTRK 372
Query: 539 VSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGK 598
FNI+SGTSMSCPH+SGLAA+LK +HP WSP+AI+SALMTT+Y + ++D A G
Sbjct: 373 TKFNIMSGTSMSCPHISGLAALLKAAHPTWSPSAIKSALMTTAYNHDNSKSPLRDAADGS 552
Query: 599 PATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCDPKKK 658
+TPL GAGHV+P +L PGLVYDA+ DY+ FLC+LNY S +I+L +R KK
Sbjct: 553 FSTPLAHGAGHVNPQKALSPGLVYDASTKDYITFLCSLNYNSEQIQLIVKRPSVNCTKKF 732
Query: 659 YRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKASVSSQSPSVKIA 718
NYPSF+V + V+Y+R +TNVG G+ V SV I
Sbjct: 733 ANPGQLNYPSFSVVFSSKR----------VVRYTRIVTNVGEAGSVYNVVVDVPSSVGIT 882
Query: 719 VEPQILRFQELYEKKSYTVTFTSNSMPSGTK---SFAYLYWSDGKHRVASPIAITWT 772
V+P L F+++ E+K YTVTF S +K F + WS+ +H+V SPIA WT
Sbjct: 883 VKPSRLVFEKVGERKRYTVTFVSKKGADASKVRSGFGSILWSNAQHQVRSPIAFAWT 1053
>TC89220 similar to PIR|T05768|T05768 subtilisin-like proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (39%)
Length = 1149
Score = 320 bits (820), Expect = 1e-87
Identities = 167/357 (46%), Positives = 228/357 (63%), Gaps = 10/357 (2%)
Frame = +3
Query: 7 QLLQSALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFDSSLQSVS 66
Q+L S + L F S + ++ KT+I ++ + P+ F H HW+ + +
Sbjct: 48 QILNSIVFFLFFFSLSLNQV-LSSDEELPKTFIFRVNSYSKPSIFPTHYHWYTAEF---T 215
Query: 67 ESAEILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLKKT 126
E IL+TY HGFS LT Q+ +++ P +L+V + R +LHTTR+P+FLGL +
Sbjct: 216 EQTNILHTYDTAFHGFSAVLTRQQVASISNHPSILAVFEDRRRQLHTTRSPQFLGLRNQR 395
Query: 127 TTLSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKL 186
S SD S V++GV DTG+WPE +S D L P+P WKG CE+G + +CNRKL
Sbjct: 396 GLWSE-SDYGSDVIVGVFDTGIWPERRSFSDMNLGPIPRRWKGVCESGEKFSPRNCNRKL 572
Query: 187 IGARFFSKGYE------ATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLA 240
IGAR+FSKG+E L PI+ + E RS RD DGHG+HT +TAAG A++ G A
Sbjct: 573 IGARYFSKGHEVGAGSAGPLNPINETVEFRSPRDADGHGTHTASTAAGRYAFQANMSGYA 752
Query: 241 SGTARGMATQARVAAYKVCWL-GGCFSSDIAAGIDKAIEDGVNIISMSIGGS---SADYF 296
SG A+G+A +AR+A YKVCW GCF SDI A D A+ DGV++IS+SIGG ++ Y+
Sbjct: 753 SGIAKGVAPKARLAVYKVCWKNSGCFDSDILAAFDAAVNDGVDVISISIGGGDGIASPYY 932
Query: 297 RDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITL 353
D IAIG++ A S G+ VS+SAGN GPS S++N APW+TTVGAGTIDRDFP+ I +
Sbjct: 933 LDPIAIGSYGAVSRGVFVSSSAGNDGPSGMSVTNLAPWLTTVGAGTIDRDFPSQIII 1103
>BG586808 weakly similar to PIR|T07171|T07 subtilisin-like proteinase (EC
3.4.21.-) 1 - tomato, partial (23%)
Length = 673
Score = 172 bits (437), Expect(2) = 9e-87
Identities = 89/135 (65%), Positives = 109/135 (79%), Gaps = 1/135 (0%)
Frame = +2
Query: 5 IFQLLQSA-LLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFDSSLQ 63
+ Q+L+S ++LLLIFCS + A Q K TYIIHMDKSTMP TF DH +WFD+SL+
Sbjct: 20 MMQILKSLQIVLLLIFCSRHITA------QTKNTYIIHMDKSTMPETFTDHLNWFDTSLK 181
Query: 64 SVSESAEILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLL 123
SVSE+AEILYTYKH+AHG+STRLT QEAETL++QPG+L V PE+RY+LHTTRTP+FLG L
Sbjct: 182 SVSETAEILYTYKHIAHGYSTRLTNQEAETLSKQPGILDVIPELRYQLHTTRTPQFLG-L 358
Query: 124 KKTTTLSPGSDKQSQ 138
KT TL P S +QS+
Sbjct: 359 PKTNTLLPHSRQQSK 403
Score = 167 bits (422), Expect(2) = 9e-87
Identities = 75/90 (83%), Positives = 82/90 (90%)
Frame = +1
Query: 139 VVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGARFFSKGYEA 198
V+IG+LDTG+WPELKSLDDTGL P+PS WKG CE GNNMNSS CN+KLIGARFF KGYEA
Sbjct: 403 VIIGILDTGIWPELKSLDDTGLGPIPSNWKGVCETGNNMNSSHCNKKLIGARFFLKGYEA 582
Query: 199 TLGPIDVSTESRSARDDDGHGSHTLTTAAG 228
LGPID +TES+SARDDDGHGSHTLTTAAG
Sbjct: 583 ALGPIDETTESKSARDDDGHGSHTLTTAAG 672
>TC89137 similar to GP|4200338|emb|CAA76726.1 P69C protein {Lycopersicon
esculentum}, partial (37%)
Length = 1230
Score = 280 bits (715), Expect = 2e-75
Identities = 165/400 (41%), Positives = 231/400 (57%), Gaps = 6/400 (1%)
Frame = +3
Query: 378 VYAG--NASNFSVGYLCLPDSLVPSKVLGKIVICERGGNA-RVEKGLVVKRAGGIGMILA 434
VYAG N S+ S+ + C P ++ V GKIV+CE+GG RV KG VK AGG MIL
Sbjct: 3 VYAGSINTSDDSIAF-CGPIAMKKVDVKGKIVVCEQGGFVGRVAKGQAVKDAGGAAMILL 179
Query: 435 NNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAA 494
N+E +AD H+LPA + + ++DY+ S+ P A ++F GT + +P VA+
Sbjct: 180 NSEGEDFNPIADVHVLPAVHVSYSAGLNIQDYINSTSTPMATILFKGTVIGNPNAPQVAS 359
Query: 495 FSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHV 554
FSSRGP+ +P ILKPD++ PG+NILAGW + +D SFNII+GTSMSCPH+
Sbjct: 360 FSSRGPSKASPGILKPDILGPGLNILAGWP-------ISLDNSTSSFNIIAGTSMSCPHL 518
Query: 555 SGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPVA 614
SG+AA+LK SHP+WSPAAI+SA+MTT+ +G+ I D PA GAGHV+P
Sbjct: 519 SGIAALLKNSHPDWSPAAIKSAIMTTANHVNLHGKPILDQRL-LPADVFATGAGHVNPSK 695
Query: 615 SLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCDPKKKYRVEDFNYPSFAVPLE 674
+ DPGLVYD +DY+ +LC LNYT +++ + ++ KC K NYPS ++ L
Sbjct: 696 ANDPGLVYDIETNDYVPYLCGLNYTDIQVGIILQQKVKCSDVKSIPQAQLNYPSISIRLG 875
Query: 675 TASGIGGGSHAPITVKYSRTLTNVGTPGTYKASVSSQSPSVKIAVEPQILRFQELYEKKS 734
S YSRTLTNVG T V +V+++V P + F E+ +K +
Sbjct: 876 NTSQF-----------YSRTLTNVGPVNTTYNVVIDVPVAVRMSVRPSQITFTEVKQKVT 1022
Query: 735 YTVTFTSNSMPSGTKSF---AYLYWSDGKHRVASPIAITW 771
Y V F + +F + W K+ V+ PIA+ +
Sbjct: 1023YWVDFIPEDKENRGDNFIAQGSIKWISAKYSVSIPIAVVF 1142
>TC81705 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428(E30359)
corresponds to a region of the predicted gene.~Similar
to antifreeze-like, partial (33%)
Length = 1443
Score = 264 bits (674), Expect = 1e-70
Identities = 151/366 (41%), Positives = 207/366 (56%), Gaps = 13/366 (3%)
Frame = +1
Query: 12 ALLLLLIFCSSYTIAEKKTAQQAKKTYIIHM------DKSTMPATFNDHQHWFDSSLQSV 65
A+ LL+ +S I A Q K Y+++M + +P + H S + S
Sbjct: 7 AMKSLLLLLASLLICNTTIADQITKPYVVYMGNNINGEDDQIPESV--HIELLSSIIPSE 180
Query: 66 -SESAEILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGL-- 122
SE ++++ Y H GFS LT EA L+ GV+SV P+ ELHTTR+ +FL
Sbjct: 181 ESERIKLIHHYNHAFSGFSAMLTQSEASALSGHDGVVSVFPDPILELHTTRSWDFLDSDL 360
Query: 123 -LKKTTTLSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSS 181
+K +T + + ++I ++DTG+WPE S D G+ +PS WKG C G++ S+
Sbjct: 361 GMKPSTNVLTHQHSSNDIIIALIDTGIWPESPSFTDEGIGKIPSVWKGICMEGHDFKKSN 540
Query: 182 CNRKLIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLAS 241
CNRKLIGAR+++ + S RD GHG+HT +TAAG V A+ +GLA
Sbjct: 541 CNRKLIGARYYNTQDTFGSNKTHIGGAKGSPRDTVGHGTHTASTAAGVNVNNANYYGLAK 720
Query: 242 GTARGMATQARVAAYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGGSS---ADYFRD 298
GTARG + R+AAYK C GC S I +D AI+DGV+IIS+SIG SS +DY D
Sbjct: 721 GTARGGSPSTRIAAYKTCSEEGCSGSTILKAMDDAIKDGVDIISISIGLSSLMQSDYLND 900
Query: 299 IIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNIT 358
IAIGAF A G+ SAGN GP P+++ NTAPWI TV A IDR+F + I LGN +
Sbjct: 901 PIAIGAFHAEQRGVTAVCSAGNDGPDPNTVVNTAPWIFTVAASNIDRNFQSTIVLGNGKS 1080
Query: 359 HTGASL 364
GA +
Sbjct: 1081FQGAGI 1098
>TC81650 similar to PIR|T07171|T07171 subtilisin-like proteinase (EC
3.4.21.-) 1 - tomato, partial (19%)
Length = 972
Score = 251 bits (640), Expect = 1e-66
Identities = 133/268 (49%), Positives = 175/268 (64%), Gaps = 5/268 (1%)
Frame = +2
Query: 510 PDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWS 569
PD+IAPGVNILAGWTG +GPT L +D R V FNIISGTSMSCPHVSG+AA+L+ ++PEWS
Sbjct: 2 PDVIAPGVNILAGWTGKVGPTDLEIDPRRVEFNIISGTSMSCPHVSGIAALLRKAYPEWS 181
Query: 570 PAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDY 629
PAAI+SALMTT+Y +G I+D+ TGK + P GAGHVDP +L+PGLVYD N++DY
Sbjct: 182 PAAIKSALMTTAYNVDNSGGKIKDLGTGKESNPFVHGAGHVDPNKALNPGLVYDLNINDY 361
Query: 630 LGFLCALNYTSLEIKLASRRDFK---CDPKKKYRVE-DFNYPSFAVPLETASGIGGGSHA 685
L FLC++ Y + EI++ +R C+ ++K+ D NYPSF+V +G+
Sbjct: 362 LAFLCSIGYDAKEIQIFTREPTSYNVCENERKFTSPGDLNYPSFSVVFGANNGL------ 523
Query: 686 PITVKYSRTLTNVGTPGTYKASVSSQSP-SVKIAVEPQILRFQELYEKKSYTVTFTSNSM 744
VKY R LTNVG +V +P V ++V P L F + +++ VTFT
Sbjct: 524 ---VKYKRVLTNVGDSVDAVYTVKVNAPFGVDVSVSPSKLVFSSENKTQAFEVTFTRIGY 694
Query: 745 PSGTKSFAYLYWSDGKHRVASPIAITWT 772
G++SF L WSDG H V SPIA W+
Sbjct: 695 -GGSQSFGSLEWSDGSHIVRSPIAARWS 775
>BI310600 similar to PIR|S52770|S52 subtilisin-like proteinase (EC 3.4.21.-)
nodule-specific - Arabidopsis thaliana (fragment),
partial (19%)
Length = 648
Score = 241 bits (616), Expect(2) = 2e-66
Identities = 117/201 (58%), Positives = 153/201 (75%), Gaps = 1/201 (0%)
Frame = +3
Query: 552 PHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVD 611
PH +AA++K HP+WSPAAIRSALMTT+YTAYKN +T+ D A KPATP DFGAGHV+
Sbjct: 9 PHAKWIAALIKSVHPDWSPAAIRSALMTTTYTAYKNNKTLLDGANKKPATPFDFGAGHVN 188
Query: 612 PVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCDPKKKYRVEDFNYPSFAV 671
P+ +L+PGLVYD VDDYL FLCALNY++ +I++ +RR + CDPKK+Y V + NYPSFAV
Sbjct: 189 PIFALNPGLVYDLTVDDYLSFLCALNYSADKIEMVARRKYTCDPKKQYSVTNLNYPSFAV 368
Query: 672 PLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKASVSSQSPSVKIAVEPQILRFQELYE 731
E G H +K++RTLTNVG GTYK S+ S +PS+KI+VEP++L F++ E
Sbjct: 369 VFE-------GEHGVEEIKHTRTLTNVGAEGTYKVSIKSDAPSIKISVEPEVLSFKK-NE 524
Query: 732 KKSYTVTFTSN-SMPSGTKSF 751
KKSY +TF+S+ S P+ T+SF
Sbjct: 525 KKSYIITFSSSGSKPNSTQSF 587
Score = 30.0 bits (66), Expect(2) = 2e-66
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = +1
Query: 749 KSFAYLYWSDGKHRVASPIAITW 771
K F L WSDGK V SPI +W
Sbjct: 580 KVFGSLEWSDGKTVVRSPIVFSW 648
>TC91562 similar to PIR|T07172|T07172 subtilisin-like proteinase (EC
3.4.21.-) 2 - tomato, partial (36%)
Length = 1062
Score = 215 bits (548), Expect(2) = 7e-65
Identities = 102/197 (51%), Positives = 137/197 (68%)
Frame = +2
Query: 475 AKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPV 534
A L F T L ++PSP+VAAFSSRGP+ LT +ILKPD++APGVNILA W+G GP+ LP+
Sbjct: 8 ATLAFHNTRLGIRPSPIVAAFSSRGPSLLTLEILKPDIVAPGVNILAAWSGLTGPSSLPI 187
Query: 535 DTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDV 594
D R V FNI+SGTSMSCPHVSG+AA++K HPEWSPAAI+SA+MTT+Y + ++D
Sbjct: 188 DHRRVKFNILSGTSMSCPHVSGIAAMIKAKHPEWSPAAIKSAIMTTAYVHDNTIKPLRDA 367
Query: 595 ATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCD 654
++ + +TP D GAGH++P +LDPGL+YD DY FLC + E+ + S+ +
Sbjct: 368 SSAEFSTPYDHGAGHINPRKALDPGLLYDIEPQDYFEFLCTKKLSPSELVVFSKNSNRNC 547
Query: 655 PKKKYRVEDFNYPSFAV 671
D NYP+ +V
Sbjct: 548 KHTLASASDLNYPAISV 598
Score = 51.2 bits (121), Expect(2) = 7e-65
Identities = 29/79 (36%), Positives = 39/79 (48%)
Frame = +1
Query: 693 RTLTNVGTPGTYKASVSSQSPSVKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKSFA 752
RT+TNVG + + + + VEP L F Y+K SY ++F S S F
Sbjct: 640 RTVTNVGPAVSKYHVIGTPFKGAVVKVEPDTLNFTRKYQKLSYKISFKVTSRQS-EPEFG 816
Query: 753 YLYWSDGKHRVASPIAITW 771
L W D H+V SPI IT+
Sbjct: 817 GLVWKDRLHKVRSPIVITY 873
>TC85586 similar to GP|20521377|dbj|BAB91889. putative subtilisin-like
protease {Oryza sativa (japonica cultivar-group)},
partial (17%)
Length = 1460
Score = 242 bits (617), Expect = 4e-64
Identities = 158/390 (40%), Positives = 216/390 (54%), Gaps = 12/390 (3%)
Frame = +1
Query: 392 CLPDSLVPSKVLGKIVICERGGNAR-VEKGLVVKRAGGIGMILANNEEF-GEELVADSHL 449
C P +L PSKV GKIV C R G + V +G AG GM L N + G L+++ H+
Sbjct: 46 CRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHV 225
Query: 450 LPAAALGERSSKALKDYVFSSRNPT----AKLVFGG--THLQVKPSPVVAAFSSRGPNGL 503
L +++ + + T K+ F T + KP+PV+A+FSSRGPN +
Sbjct: 226 LSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQV 405
Query: 504 TPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRH-VSFNIISGTSMSCPHVSGLAAILK 562
P ILKPD+ APGVNILA ++ + L D R FN++ GTSMSCPHV+G A ++K
Sbjct: 406 QPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIK 585
Query: 563 GSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAG-HVDPVASLDPGLV 621
HP WSPAAI+SA+MTT+ T + I D A P +G+G + P +++DPGLV
Sbjct: 586 TLHPNWSPAAIKSAIMTTATTRDNTNKPISDAFDKTLADPFAYGSGTYSHPTSAIDPGLV 765
Query: 622 YDANVDDYLGFLCALNYTSLEIK-LASRRDFKCDPKKKYRVEDFNYPSFAVPLETASGIG 680
YD + DYL FLCA Y I L F C + ++D NYPS +P +G
Sbjct: 766 YDLGIKDYLNFLCASGYNKQLISALNFNMTFTC--SGTHSIDDLNYPSITLP-----NLG 924
Query: 681 GGSHAPITVKYSRTLTNVGTPGTYKASVSSQSPSVKIAVEPQILRFQELYEKKSYTVTFT 740
+ ITV +RT+TNVG P TY A V Q P KIAV P L F+++ EKK++ V
Sbjct: 925 LNA---ITV--TRTVTNVGPPSTYFAKV--QLPGYKIAVVPSSLNFKKIGEKKTFQVIVQ 1083
Query: 741 SNS-MPSGTKSFAYLYWSDGKHRVASPIAI 769
+ S +P F L W++GKH V SP+ +
Sbjct: 1084ATSEIPRRKYQFGELRWTNGKHIVRSPVTV 1173
>TC85584 weakly similar to PIR|A84473|A84473 probable serine proteinase
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1241
Score = 231 bits (590), Expect = 6e-61
Identities = 146/362 (40%), Positives = 199/362 (54%), Gaps = 9/362 (2%)
Frame = +1
Query: 417 VEKGLVVKRAGGIGMILANNEEF-GEELVADSHLLPAAALGERSSKALK---DYVFSSRN 472
V +G AG G+IL N E G+ L+++ H+L + S+ D + S
Sbjct: 13 VAEGQEALSAGAKGVILRNQPEINGKTLLSEPHVLSTISYPGNHSRTTGRSLDIIPSDIK 192
Query: 473 PTAKLVFGG--THLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPT 530
KL T + KP+PV+A++SSRGPN + P ILKPD+ APGVNILA ++ +
Sbjct: 193 SGTKLRMSPAKTLNRRKPAPVMASYSSRGPNKVQPSILKPDVTAPGVNILAAYSLFASAS 372
Query: 531 GLPVDTRH-VSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQ 589
L DTR FN++ GTSMSCPHV+G A ++K HP WSPAAI+SA+MTT+ T +
Sbjct: 373 NLITDTRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATTRDNTNK 552
Query: 590 TIQDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIK-LASR 648
I D A P +G+GH+ P +++DPGLVYD + DYL FLCA Y I L
Sbjct: 553 PISDAFDKTLANPFAYGSGHIRPNSAMDPGLVYDLGIKDYLNFLCASGYNQQLISALNFN 732
Query: 649 RDFKCDPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKASV 708
F C ++D NYPS +P +G S V +RT+TNVG P TY A V
Sbjct: 733 MTFTCSGTSS--IDDLNYPSITLP-----NLGLNS-----VTVTRTVTNVGPPSTYFAKV 876
Query: 709 SSQSPSVKIAVEPQILRFQELYEKKSYTVTFTSNSM-PSGTKSFAYLYWSDGKHRVASPI 767
Q KIAV P L F+++ EKK++ V + S+ P F L W++GKH V SP+
Sbjct: 877 --QLAGYKIAVVPSSLNFKKIGEKKTFQVIVQATSVTPRRKYQFGELRWTNGKHIVRSPV 1050
Query: 768 AI 769
+
Sbjct: 1051TV 1056
>TC83163 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428(E30359)
corresponds to a region of the predicted gene.~Similar
to antifreeze-like, partial (29%)
Length = 990
Score = 229 bits (584), Expect = 3e-60
Identities = 137/341 (40%), Positives = 186/341 (54%), Gaps = 10/341 (2%)
Frame = +2
Query: 5 IFQLLQSALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFDSSLQS 64
+ Q L L L+ CSS + + Q K Y+++M + + L S
Sbjct: 2 LLQFLHLLFLASLLICSSSS--NITISDQIAKPYVVYMGSNINGVDGQIPESVHLDLLSS 175
Query: 65 V-----SESAEILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEF 119
+ SE +++ Y H +GFS LT EA LA GV+SV + ELHTTR+ +F
Sbjct: 176 IIPSEESERIALIHHYSHAFNGFSAMLTQSEASALAGNDGVVSVFEDPFLELHTTRSWDF 355
Query: 120 L--GLLKKTTTLSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNM 177
L L + + + ++IGV+DTG+WPE S D G+ +PS WKG C ++
Sbjct: 356 LESDLGMRPHGILKHQHSSNDIIIGVIDTGIWPESPSFKDEGIGKIPSRWKGVCMEAHDF 535
Query: 178 NSSSCNRKLIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLF 237
S+CNRKLIGAR+++K S RD +GHG+HT +TAAG V AS +
Sbjct: 536 KKSNCNRKLIGARYYNK-----------KDPKGSPRDFNGHGTHTASTAAGVIVNNASYY 682
Query: 238 GLASGTARGMATQARVAAYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGGSS---AD 294
GLA GTARG + AR+AAYK C GC + ID AI+DGV+IIS+SIG SS ++
Sbjct: 683 GLAKGTARGGSPSARIAAYKACSGEGCSGGTLLKAIDDAIKDGVDIISISIGFSSEFLSE 862
Query: 295 YFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWI 335
Y D IAIGAF A G++V SAGN GP ++ NT PWI
Sbjct: 863 YLSDPIAIGAFHAEQRGVMVVCSAGNEGPDHYTVVNTTPWI 985
>TC88072 similar to PIR|T06580|T06580 subtilisin-like proteinase (EC
3.4.21.-) p69f - tomato, partial (27%)
Length = 1097
Score = 228 bits (580), Expect = 9e-60
Identities = 136/329 (41%), Positives = 188/329 (56%), Gaps = 4/329 (1%)
Frame = +2
Query: 445 ADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLT 504
A +H+LPA + + +K Y+ S+ NPTA L+F GT + +P V FSSRGP+ +
Sbjct: 5 AIAHVLPAVEVSYAAGLTIKSYIKSTYNPTATLIFKGTIIGDSLAPSVVYFSSRGPSQES 184
Query: 505 PKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGS 564
P ILKPD+I PGVNILA W + VD + +F+I+SGTSMSCPH+SG+AA++K S
Sbjct: 185 PGILKPDIIGPGVNILAAWA-------VSVDNKIPAFDIVSGTSMSCPHLSGIAALIKSS 343
Query: 565 HPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPVASLDPGLVYDA 624
HP+WSPAAI+SA+MTT+ T G I D PA GAGHV+P + DPGLVYD
Sbjct: 344 HPDWSPAAIKSAIMTTANTLNLGGIPILDQRL-LPADIFATGAGHVNPFKANDPGLVYDI 520
Query: 625 NVDDYLGFLCALNYTSLEIKLASRRDFKCDPKKKYRVEDFNYPSFAVPLETASGIGGGSH 684
+DY+ +LC L Y+ EI++ + KC K NYPSF++ L + S
Sbjct: 521 EPEDYVPYLCGLGYSDKEIEVIVQWKVKCSNVKSIPEAQLNYPSFSILLGSDSQY----- 685
Query: 685 APITVKYSRTLTNVG-TPGTYKASVSSQSPSVKIAVEPQILRFQELYEKKSYTVTF---T 740
Y+RTLTNVG TYK + ++ ++V P + F E+ EK S++V F
Sbjct: 686 ------YTRTLTNVGFANSTYKVELEVPL-ALGMSVNPSEITFTEVNEKVSFSVEFIPQI 844
Query: 741 SNSMPSGTKSFAYLYWSDGKHRVASPIAI 769
+ + T L W KH V PI++
Sbjct: 845 KENRRNQTFGQGSLTWVSDKHAVRVPISV 931
>TC85585 similar to GP|22773236|gb|AAN06842.1 Putatvie subtilisin-like
serine protease {Oryza sativa (japonica
cultivar-group)}, partial (21%)
Length = 1353
Score = 224 bits (572), Expect = 7e-59
Identities = 128/286 (44%), Positives = 168/286 (57%), Gaps = 3/286 (1%)
Frame = +2
Query: 487 KPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRH-VSFNIIS 545
KP+PV+A+FSSRGPN + P ILKPD+ APGVNILA ++ + L D R FNI
Sbjct: 134 KPAPVMASFSSRGPNKVQPYILKPDVTAPGVNILAAYSLLASVSNLVTDNRRGFPFNIQQ 313
Query: 546 GTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDF 605
GTSMSCPHV G A ++K HP WSPAAI+SA+MTT+ T + I+D A +
Sbjct: 314 GTSMSCPHVVGTAGLIKTLHPNWSPAAIKSAIMTTATTRDNTNEPIEDAFENTTANAFAY 493
Query: 606 GAGHVDPVASLDPGLVYDANVDDYLGFLCALNYT-SLEIKLASRRDFKCDPKKKYRVEDF 664
G+GH+ P +++DPGLVYD + DYL FLCA Y L L F C + + D
Sbjct: 494 GSGHIQPNSAIDPGLVYDLGIKDYLNFLCAAGYNQKLISSLIFNMTFTCYGTQS--INDL 667
Query: 665 NYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKASVSSQSPSVKIAVEPQIL 724
NYPS +P +G + V +RT+TNVG TY A +Q P KI V P L
Sbjct: 668 NYPSITLP-----NLGLNA-----VTVTRTVTNVGPRSTYTA--KAQLPGYKIVVVPSSL 811
Query: 725 RFQELYEKKSYTVTFTSNSM-PSGTKSFAYLYWSDGKHRVASPIAI 769
+F+++ EKK++ VT + S+ P G F L WS+GKH V SPI +
Sbjct: 812 KFKKIGEKKTFKVTVQATSVTPQGKYEFGELQWSNGKHIVRSPITL 949
>CB892799 weakly similar to PIR|T05768|T05 subtilisin-like proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (20%)
Length = 875
Score = 216 bits (549), Expect = 3e-56
Identities = 122/290 (42%), Positives = 181/290 (62%), Gaps = 6/290 (2%)
Frame = +2
Query: 298 DIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNI 357
D I+IG+F A + G+ V +SAGN G + S +N APW+ TV AG+ DRDF + I LGN
Sbjct: 2 DAISIGSFHAANRGLFVVSSAGNEG-NLGSATNLAPWMLTVAAGSTDRDFTSDIILGNGA 178
Query: 358 THTGASLYRGKPLSDSPL---PLVYAGNASNFSVGYLCLPDSLVPSKVLGKIVIC---ER 411
TG SL + + + + +AG + + Y CL SL +K GK+++C ER
Sbjct: 179 KITGESLSLFEMNASTRIISASEAFAGYFTPYQSSY-CLESSLNKTKTKGKVLVCRHVER 355
Query: 412 GGNARVEKGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSR 471
++V K +VK AGG+GMIL + + + VA ++P+A +G++ + + Y+ ++R
Sbjct: 356 STESKVAKSKIVKEAGGVGMILIDETD---QDVAIPFVIPSAIVGKKKGQKILSYLKTTR 526
Query: 472 NPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTG 531
P +K++ T + + +P VAAFSSRGPN L P+ILKPD+ APG+NILA W+ G
Sbjct: 527 KPMSKILRAKTVIGAQSAPRVAAFSSRGPNALNPEILKPDITAPGLNILAAWSPVAGN-- 700
Query: 532 LPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTS 581
FNI+SGTSM+CPHV+G+A ++K HP WSP+AI+SA+MTT+
Sbjct: 701 --------MFNILSGTSMACPHVTGIATLVKAVHPSWSPSAIKSAIMTTA 826
>BI307979 weakly similar to GP|20521377|db putative subtilisin-like protease
{Oryza sativa (japonica cultivar-group)}, partial (18%)
Length = 777
Score = 214 bits (545), Expect = 1e-55
Identities = 121/256 (47%), Positives = 161/256 (62%), Gaps = 5/256 (1%)
Frame = +3
Query: 273 IDKAIEDGVNIISMSIG-GSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNT 331
ID AIEDGV++I++SIG + Y D+IA GA A I+V SAGN GPSP SLSN
Sbjct: 12 IDDAIEDGVDVINLSIGFPAPLKYEDDVIAKGALQAVRKNIVVVCSAGNAGPSPHSLSNP 191
Query: 332 APWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYAGNASNFSV--- 388
+PWI TVGA T+DR F A I L N T G S+ + + +S PLV A + +
Sbjct: 192 SPWIITVGASTVDRTFLAPIKLSNGTTIEGRSITPLR-MGNSFCPLVLASDVEYAGILSA 368
Query: 389 -GYLCLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEELVADS 447
CL ++L PSKV GKIV+C RG RV+K L V+RAGG+G+IL NN+ + ++ +D
Sbjct: 369 NSSYCLDNTLDPSKVKGKIVLCMRGQGGRVKKSLEVQRAGGVGLILGNNKVYANDVPSDP 548
Query: 448 HLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKI 507
+ +P + ++ L Y+ SS NP A+L+ G T L KP+P +A FSSRGPN + P I
Sbjct: 549 YFIPTTGVTYENTLKLVQYIHSSPNPMAQLLPGRTVLDTKPAPSMAIFSSRGPNIIDPNI 728
Query: 508 LKPDLIAPGVNILAGW 523
LKPD+ APGV+I A W
Sbjct: 729 LKPDITAPGVDIFAAW 776
>CA990312 weakly similar to GP|14488360|gb putative serine protease {Oryza
sativa (japonica cultivar-group)}, partial (20%)
Length = 574
Score = 214 bits (544), Expect = 1e-55
Identities = 106/183 (57%), Positives = 135/183 (72%), Gaps = 1/183 (0%)
Frame = +3
Query: 298 DIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNI 357
D +AIGAF A HGILVS +AGN GP+P S++N APWITTVGAGT+DRDFPAYI+LGN
Sbjct: 21 DNLAIGAFAAMEHGILVSCAAGNSGPNPLSVTNVAPWITTVGAGTLDRDFPAYISLGNGK 200
Query: 358 THTGASLYRGKPLSDSPLPLVYAGNASNFSVGY-LCLPDSLVPSKVLGKIVICERGGNAR 416
+ G SL +G L D+P+P +YAGNAS +G C+ SL P KV GKIV+C+RG ++R
Sbjct: 201 KYPGVSLSKGNSLPDTPVPFIYAGNASINGLGTGTCISGSLDPKKVSGKIVLCDRGESSR 380
Query: 417 VEKGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAK 476
EKG VK AGG+GM+LAN E GEE VAD+H+LPA A+G + +A+K Y+F PTA
Sbjct: 381 TEKGNTVKSAGGLGMVLANVESDGEEPVADAHILPATAVGFKDGEAIKKYLFFDPKPTAT 560
Query: 477 LVF 479
++F
Sbjct: 561 ILF 569
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,881,896
Number of Sequences: 36976
Number of extensions: 294395
Number of successful extensions: 1752
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 1594
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1605
length of query: 772
length of database: 9,014,727
effective HSP length: 104
effective length of query: 668
effective length of database: 5,169,223
effective search space: 3453040964
effective search space used: 3453040964
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0096b.8


