

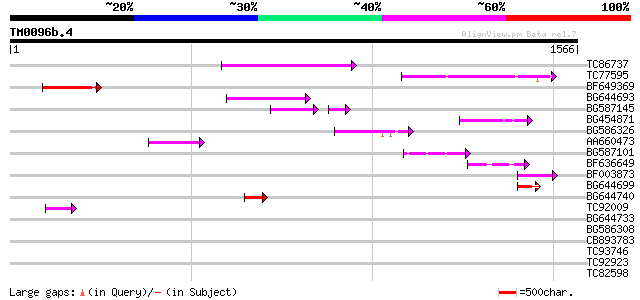
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0096b.4
(1566 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 245 9e-65
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 192 7e-49
BF649369 172 9e-43
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 159 6e-39
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 105 3e-31
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 126 8e-29
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 98 2e-20
AA660473 75 2e-13
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 72 1e-12
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 69 1e-11
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 60 9e-09
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 59 1e-08
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 58 3e-08
TC92009 47 8e-05
BG644733 weakly similar to GP|15289942|db putative polyprotein {... 41 0.003
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 40 0.005
CB893783 weakly similar to GP|22830935|dbj hypothetical protein~... 39 0.012
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 36 0.13
TC92923 similar to GP|6466937|gb|AAF13073.1| putative retroeleme... 35 0.30
TC82598 weakly similar to PIR|C84547|C84547 hypothetical protein... 32 1.9
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 245 bits (626), Expect = 9e-65
Identities = 145/390 (37%), Positives = 220/390 (56%), Gaps = 16/390 (4%)
Frame = +1
Query: 585 ILERHSVVFQAPKG--LPPKRNK-QHAITL---KEG-EGPVNVRP-YRYPHHQKNEIENQ 636
+LE +F K +P R HAI L K+G + P+ P Y + ++
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 637 VKELLEGGVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLDE 696
+++LL+ G I+ S S+ +PV+ V+K R CVDYRALN T D++P+P+I E L
Sbjct: 523 LEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRR 702
Query: 697 LHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMND 756
+ GAR+F+KLD+ + +H++R+K+ED KTAFRT G +E++V PFGL AP+TFQ +N
Sbjct: 703 VAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINK 882
Query: 757 IFRHLLRKRVLVFFDDILVYSK-DWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEY 815
L V + DD+L+Y+ H ++ VL L + GL + KKC F V+Y
Sbjct: 883 TLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKY 1062
Query: 816 LGHMI-SGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTEL 874
+G ++ +G+GV DP K+ ++ W P +VKG R FLG YY+ FI Y +I +PLT L
Sbjct: 1063VGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRL 1242
Query: 875 TKKN-GFEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQE--- 930
T+K+ F W + + AF LK+ PVL + D T+E D SG +G +L QE
Sbjct: 1243TRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDGT 1422
Query: 931 --KRPIAYFSKALGVRNLSKSAYEKELMAL 958
P+A+ S+ L + ++KEL+A+
Sbjct: 1423GAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 192 bits (489), Expect = 7e-49
Identities = 142/448 (31%), Positives = 225/448 (49%), Gaps = 21/448 (4%)
Frame = +2
Query: 1082 LLYEGRLVLP----RESPL--IHTMLT-EFHTTPQGGHSGFYRTYRRLAANVYWRGMKSA 1134
L + GR+ +P ESPL + T L E H + GH G T ++ +W G
Sbjct: 104 LTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQT 283
Query: 1135 VQDFVKQCDVCQRQKYLASSPGGLLQPLPIPERIWEDLSMDFITGLP--KSKGFEAILVV 1192
V+ FV+ CDVC + G L+PLP+P R+ DLSMDFIT LP + +G + + V+
Sbjct: 284 VRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVI 463
Query: 1193 VDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELFKLQ 1252
VDRLSK + A++ A+ F R HG+P SIVSDR +V FWRE +L
Sbjct: 464 VDRLSKSVTLEEMD-TMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLT 640
Query: 1253 GTKLKMSTAYHPESDGQSEVVNRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHASTG 1312
G +ST+YHP++DG +E N+ ++ LR ++ W +P + ++S G
Sbjct: 641 GVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIG 820
Query: 1313 VTPFEVVYG---RPPPTITRWIQGETRVEAVQKELLER-DEALRQLRLQLARAQDRMKQF 1368
TPF V +G P PT+ + EA + L++R + ++ ++ AQ R +
Sbjct: 821 ATPFFVEHGYHVDPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEAS 1000
Query: 1369 ADRKR--SDRSFSIGEWVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLK 1426
A+++R +DR + +G+ V++ + ++ +K + Y V V +L
Sbjct: 1001 ANKRRCPADR-YQVGDKVWLNVSNYKSPRP-----SKKLDWLHHKYEVTRFVTPHVVELN 1162
Query: 1427 LPPGSKVHPVFHVSLLKKAVGTYHEGEEL------PDLEGDGGILIEPTEVLATRTVQLQ 1480
+P V+P FHV LL++A G+E+ P ++ DG + E E+LA R Q+
Sbjct: 1163 VP--GTVYPKFHVDLLRRAASDPLPGQEVVDPQPPPIVDDDGEVEWEVEEILAARWHQVG 1336
Query: 1481 GQSIKQILIQWKGQQPEEATWEDVDMIK 1508
+Q L++WKG +ATWE D I+
Sbjct: 1337 RGRRRQALVKWKGF--VDATWEAADAIR 1414
>BF649369
Length = 631
Score = 172 bits (436), Expect = 9e-43
Identities = 84/163 (51%), Positives = 124/163 (75%)
Frame = +3
Query: 90 SEFRQSAKKVELPMFDGDDPAGWISRAEVYFRVQDTPPEVRASLAQLCMEGPTIHFFNSL 149
+E R + KKV+LP+F+GDDP WI+RAE+YF VQ+TP ++R L++L MEGPTIH+FN L
Sbjct: 99 NESRLAGKKVKLPLFEGDDPVAWITRAEIYFDVQNTPDDMRVKLSRLSMEGPTIHWFNLL 278
Query: 150 LSEEENLTWERFKCALLERYGGQGDGDVYEQLTELRQRGTVEEYITAFEYLTAQIPRLPE 209
+ E++L+ E+ K AL+ RY G+ + +E+L+ LRQ G+VEE++ AFE L++Q+ RLPE
Sbjct: 279 METEDDLSREKLKKALIARYDGRRLENPFEELSTLRQIGSVEEFVEAFELLSSQVGRLPE 458
Query: 210 KQFLGYFLHGLKGEIRGRVRSMVTMADLSRMKILQIARAVERE 252
+Q+LGYF+ GLK IR RVR T+ +RM++++IA+ VE E
Sbjct: 459 EQYLGYFMSGLKAHIRRRVR---TLNPTTRMQMMRIAKDVEDE 578
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 159 bits (403), Expect = 6e-39
Identities = 94/233 (40%), Positives = 133/233 (56%)
Frame = +2
Query: 599 LPPKRNKQHAITLKEGEGPVNVRPYRYPHHQKNEIENQVKELLEGGVIRHSTSSFSSPVI 658
+PP+ I L P+ + YR + ++ Q+K+LLE G I+ S V+
Sbjct: 17 VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVL 196
Query: 659 LVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLDELHGARYFSKLDLKSGYHQVRVK 718
+KKKD RM +DY LN I K+P+P+I+EL D L G+++F K+DL+ G HQ RV
Sbjct: 197 FLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVI 376
Query: 719 EEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLRKRVLVFFDDILVYSK 778
EDV KTAFR GHYE LVM FG N P F LMN +F+ L V+VF +DIL+YSK
Sbjct: 377 GEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSK 556
Query: 779 DWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYLGHMISGQGVEVDPSK 831
+ H HL+ L +L++ GL + E + H+ISG+G++VD +
Sbjct: 557 NENEHENHLRLALKVLKDIGLCQISYV*IL-VEVGFFSLHVISGEGLKVDSKR 712
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 105 bits (261), Expect(2) = 3e-31
Identities = 55/133 (41%), Positives = 80/133 (59%)
Frame = +2
Query: 720 EDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLRKRVLVFFDDILVYSKD 779
+D+ KTAF T G Y + VMPFGL NA ST+Q L+N +F L + V+ DD+LV S
Sbjct: 11 DDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSLR 190
Query: 780 WPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYLGHMISGQGVEVDPSKVESVTSWP 839
HL HL+E L E + N KC FG E+LG++++ QG+EV+P ++ ++ P
Sbjct: 191 ATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLP 370
Query: 840 TPKNVKGVRGFLG 852
+PKN + V+ G
Sbjct: 371 SPKNSREVQRLTG 409
Score = 50.4 bits (119), Expect(2) = 3e-31
Identities = 27/66 (40%), Positives = 38/66 (56%), Gaps = 4/66 (6%)
Frame = +3
Query: 880 FEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQEKR----PIA 935
F W EK +EAFE LK+ LTT PVL+ P+ ++ S V ++L++E R PI
Sbjct: 495 FVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIAISSTAVSSVLIREDRGEQKPIF 674
Query: 936 YFSKAL 941
Y SK +
Sbjct: 675 YTSKRM 692
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 126 bits (316), Expect = 8e-29
Identities = 75/203 (36%), Positives = 106/203 (51%), Gaps = 1/203 (0%)
Frame = +2
Query: 1242 SNFWRELFKLQGTKLKMSTAYHPESDGQSEVVNRCLETYLRCFIADQPKTWVIWIPWAEY 1301
SNFW++LFKL GT L MS+AYHP SDGQSE +N+ E YLRC + P W PWAEY
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1302 WYNTCFHASTGVTPFEVVYGRPPPTITRWIQGETRVEAVQKELLERDEALRQLRLQLARA 1361
WYNT ++ S +TPF+ +YGR + R +Q +L +R+E L QL+ R
Sbjct: 212 WYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQREELLSQLQSISTRL 391
Query: 1362 QDRMKQFADRKRSDRSFSIGEWVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARV-GA 1420
K + + + + WV L +++ + +P + +
Sbjct: 392 N---KL*SIKLIRNAAILSSSWVSTSL---*NCNLINSLR*HCGNIKSSVHPTLVHY*QS 553
Query: 1421 VAYQLKLPPGSKVHPVFHVSLLK 1443
AY+L LP +KV P+FHVS LK
Sbjct: 554 AAYKLSLPSTAKVPPIFHVSQLK 622
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 98.2 bits (243), Expect = 2e-20
Identities = 66/242 (27%), Positives = 117/242 (48%), Gaps = 25/242 (10%)
Frame = +2
Query: 898 TTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIAYFSKALGVRNLSKSAYEKELMA 957
T++P+L LP+ + + DAS G+G +L Q ++ IAY S+ L + ++ E+ A
Sbjct: 8 TSAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 958 LGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQKVVTMEQQNWAAKLLGFDFEISYKPGKL 1017
+ A++ WR YL G ++ TD +SLK + Q + + Q+ W + +D +I+Y PGK
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 1018 NKGADALSR----------------VNETLELRQMGSHVDWLGGKDLKE---------EV 1052
N ADALSR + L L + + LG + + +
Sbjct: 365 NLVADALSRRRVDVSAEREADDLDGMVRALRLNVLTKATESLGLEAVNQADLFTRIRLAQ 544
Query: 1053 SKDEELQRIIKSVHEKKDSSLGYTYENGILLYEGRLVLPRESPLIHTMLTEFHTTPQGGH 1112
+DE LQ++ ++ D + T ++G +L GR+ +P + L +++E H + H
Sbjct: 545 GQDENLQKVAQN-----DRTEYQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKSRFSVH 709
Query: 1113 SG 1114
G
Sbjct: 710 PG 715
>AA660473
Length = 655
Score = 75.1 bits (183), Expect = 2e-13
Identities = 56/160 (35%), Positives = 85/160 (53%), Gaps = 6/160 (3%)
Frame = -2
Query: 383 LAVEVIEEEESSEGELSSMSLSQVEQVGK---DKPQTIKLLGLIQGLPIVILIDSGATHN 439
+ +E+ + E+ +SM L ++ + K KP T++L+G ++G+PIVIL+D GA HN
Sbjct: 651 MTIELKDGEKEDILVYNSMGLCEMTEFNKLKVVKPATLQLVGCLKGVPIVILVDIGANHN 472
Query: 440 FVSTSLVHKLGKTVVDTPSLRITLGDGSQARTKGKCK-ELMIIAGNHPLCVDAQLFELGN 498
F S V +G S +G S+ G + G H VDA ELG
Sbjct: 471 FDFASSVSDVGCKGRVILSQESEVG*WSENINVG*AY*SRGTVRGFHC*GVDAYELELGE 292
Query: 499 VDMVLGIEWLRTLGD-MIVNWDKKTMSF-WSGHKWVTLQG 536
DM LG+ WL LG+ ++ +WD++T+ F W G + V LQG
Sbjct: 291 FDMFLGVAWLEKLGN*VVFDWDERTICFEWKG-EVVKLQG 175
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 72.4 bits (176), Expect = 1e-12
Identities = 54/186 (29%), Positives = 86/186 (46%), Gaps = 1/186 (0%)
Frame = +2
Query: 1087 RLVLPRESPLIHTMLTEFHTTPQGGHSGFYRTYRRLA-ANVYWRGMKSAVQDFVKQCDVC 1145
R V E P I L H + GH +T ++ A +W M F+ +CD C
Sbjct: 74 RCVAEEEIPGI---LFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPC 244
Query: 1146 QRQKYLASSPGGLLQPLPIPERIWEDLSMDFITGLPKSKGFEAILVVVDRLSKYAHFIPL 1205
QRQ + S + Q + +++ +DF+ P S + ILV VD +SK+ I
Sbjct: 245 QRQGNI-S*RNEMPQNFILEVEVFDVWGIDFMGPFPSSYNNKYILVAVDYVSKWVEAIA- 418
Query: 1206 KHPYTAKSVAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPE 1265
A V ++F I GVP ++SD F++ + +L K G + K++TAYHP+
Sbjct: 419 SPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
Query: 1266 SDGQSE 1271
+S+
Sbjct: 599 KAERSK 616
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 68.9 bits (167), Expect = 1e-11
Identities = 52/177 (29%), Positives = 87/177 (48%), Gaps = 7/177 (3%)
Frame = +1
Query: 1265 ESDGQSEVVNRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHASTGVTPFEVVYGRPP 1324
+SD Q+ ++N LET+L F ++Q ++ WAE YNT FH + G TPF+VVY
Sbjct: 4 DSDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY---- 171
Query: 1325 PTITRWIQGETRVEAVQKELLERDEALRQLRL-----QLARAQDRMKQFADRK--RSDRS 1377
+ + V ++L+ R+E L + + RA + + + A R
Sbjct: 172 ------VVAHLQKFVVARDLIYRNEGLHKSST*TSFGRGTRAYEALSRPAYETC*HPRRP 333
Query: 1378 FSIGEWVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKLPPGSKVH 1434
SI V+ R ++ ++ K A YGPY V+ ++G+VA++L LP ++H
Sbjct: 334 LSI---VYT-----RDRTYEWQVLPKYVA*CYGPYQVIKQIGSVAFKL*LPEQHQIH 480
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 59.7 bits (143), Expect = 9e-09
Identities = 35/113 (30%), Positives = 55/113 (47%), Gaps = 3/113 (2%)
Frame = +2
Query: 1403 KLAAKYYGPYPVVARVGAVAYQLKLPPG-SKVHPVFHVSLLKKAVGTYHEGEELPDLEGD 1461
KL ++ GPY + RVG VAY++ LPP +H VFHVS L+K V + D++
Sbjct: 29 KLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHVIQSDDVQVR 208
Query: 1462 GGILIE--PTEVLATRTVQLQGQSIKQILIQWKGQQPEEATWEDVDMIKSQFP 1512
+ +E P + + L+G+ I + + W E TWE + +P
Sbjct: 209 DNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVESYP 367
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 59.3 bits (142), Expect = 1e-08
Identities = 32/65 (49%), Positives = 41/65 (62%), Gaps = 1/65 (1%)
Frame = +2
Query: 1403 KLAAKYYGPYPVVARVGAVAYQLKLPPG-SKVHPVFHVSLLKKAVGTYHEGEELPDLEGD 1461
KL+ +Y GP+ V+ R+G VAY+L LPPG S VHPVFHVS+ K+ YH GD
Sbjct: 65 KLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVSMFKR----YH---------GD 205
Query: 1462 GGILI 1466
G +I
Sbjct: 206 GNYII 220
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 57.8 bits (138), Expect = 3e-08
Identities = 26/63 (41%), Positives = 40/63 (63%)
Frame = -1
Query: 649 STSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLDELHGARYFSKLDL 708
S S + ++ V+KKD +RMC+DYR NK T +K+P+P I+ L D++ YF +DL
Sbjct: 256 SISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NIDL 77
Query: 709 KSG 711
+ G
Sbjct: 76 RLG 68
>TC92009
Length = 974
Score = 46.6 bits (109), Expect = 8e-05
Identities = 25/87 (28%), Positives = 44/87 (49%), Gaps = 1/87 (1%)
Frame = +2
Query: 98 KVELPMFDGDDPAGWISRAEVYFRVQDTPPEVRASLAQLCMEG-PTIHFFNSLLSEEENL 156
K +L F G DPAGWI+RAE++F + + A + ME + +F S E +
Sbjct: 680 KYKLSQFTGTDPAGWITRAEMFFADNEIHSCDKLQWAFMSMEDEKALLWFYSWCQENPDA 859
Query: 157 TWERFKCALLERYGGQGDGDVYEQLTE 183
W+ F A++ +G + + +Q+ +
Sbjct: 860 DWKSFSMAMIREFGARMEHRTDKQMVQ 940
>BG644733 weakly similar to GP|15289942|db putative polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 174
Score = 41.2 bits (95), Expect = 0.003
Identities = 22/43 (51%), Positives = 28/43 (64%)
Frame = -3
Query: 1184 KGFEAILVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLH 1226
K +E+I VVVDRL+K FIP K Y+AK A + EIV +H
Sbjct: 130 KSYESI*VVVDRLTKSTLFIPFKTSYSAK*YARILLDEIVCIH 2
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 40.4 bits (93), Expect = 0.005
Identities = 18/52 (34%), Positives = 32/52 (60%)
Frame = -2
Query: 1226 HGVPSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPESDGQSEVVNRCL 1277
HG+P IV+D F+SN +RE + +L ++ +P+S+GQ+E N+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKII 530
>CB893783 weakly similar to GP|22830935|dbj hypothetical protein~similar to
gag-pol polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 853
Score = 39.3 bits (90), Expect = 0.012
Identities = 51/213 (23%), Positives = 81/213 (37%), Gaps = 21/213 (9%)
Frame = +2
Query: 423 IQGLPIVILIDSGATHNFVSTSLVHKLGKTVVDTPSLRIT--LGDGSQARTKGKCKELMI 480
+QG+ ++IDSG+ N VS +V KL D P L G++ R C
Sbjct: 245 LQGIVCDVIIDSGSCENVVSNYMVEKLELPTKDHPHRYKLQWLKKGNEVRVSKCCLVSFS 424
Query: 481 IAGNHPLCVDAQLFELGNVDMVLGIEWLRTLGDMIVNWDKKTMSFWSGH----------- 529
I + V + + M+LG W +D+ + + GH
Sbjct: 425 IGQKYKDNVWCDVISMDACHMLLGRPW---------QYDRHAL--YDGHANTYTFVKYGV 571
Query: 530 --KWVTLQGHEEQEGL--LVALQTMISRAGF----SGYLGKEKVQLEKDNKGVTGVQQAE 581
K V L + EG + +++S+ F + L K N+ T ++ E
Sbjct: 572 KIKLVPLPPNAFDEGKKDFKPIVSLVSKEPFKVTTKDIQDMSLILLVKSNEESTIQKEVE 751
Query: 582 LDMILERHSVVFQAPKGLPPKRNKQHAITLKEG 614
++ V + P GLPP R+ QHAI G
Sbjct: 752 HLLVDFTDVVPSEIPSGLPPMRDIQHAIDFIPG 850
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 35.8 bits (81), Expect = 0.13
Identities = 20/47 (42%), Positives = 32/47 (67%), Gaps = 1/47 (2%)
Frame = +2
Query: 1158 LLQPLPIPERIWEDLSMDFITGLPKSKGF-EAILVVVDRLSKYAHFI 1203
LL+P+P P WED+++DF GL ++ ++ +VV D+ S+ AHFI
Sbjct: 464 LLRPVPKPP--WEDVTIDFSLGLL*TQQLKDSKMVVGDKFSRMAHFI 598
>TC92923 similar to GP|6466937|gb|AAF13073.1| putative retroelement pol
polyprotein {Arabidopsis thaliana}, partial (1%)
Length = 625
Score = 34.7 bits (78), Expect = 0.30
Identities = 24/72 (33%), Positives = 41/72 (56%), Gaps = 1/72 (1%)
Frame = +2
Query: 1443 KKAVGTYHEGEELPD-LEGDGGILIEPTEVLATRTVQLQGQSIKQILIQWKGQQPEEATW 1501
K +VG+Y +LP+ +E + +P +LATR ++ +G I+WK + E A+
Sbjct: 266 KSSVGSYPVELQLPEGMEVEINDEADPQFMLATRQIR-EGDITT**AIKWKAKTLEHASL 442
Query: 1502 EDVDMIKSQFPS 1513
+D I+SQFP+
Sbjct: 443 KDDFTIRSQFPT 478
>TC82598 weakly similar to PIR|C84547|C84547 hypothetical protein At2g17030
[imported] - Arabidopsis thaliana, partial (2%)
Length = 1050
Score = 32.0 bits (71), Expect = 1.9
Identities = 12/33 (36%), Positives = 22/33 (66%)
Frame = +2
Query: 1354 LRLQLARAQDRMKQFADRKRSDRSFSIGEWVFV 1386
L+L++ R +R K++ + +R F +GEW+FV
Sbjct: 923 LKLKVFRLNERKKEWRSKSLRNRGFVLGEWMFV 1021
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,786,715
Number of Sequences: 36976
Number of extensions: 638560
Number of successful extensions: 3202
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 3122
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3181
length of query: 1566
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1457
effective length of database: 4,984,343
effective search space: 7262187751
effective search space used: 7262187751
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0096b.4


