

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0096a.2
(239 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
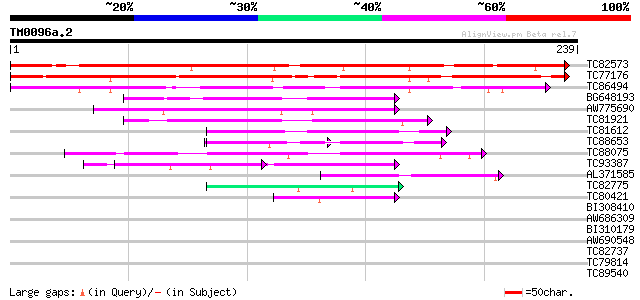
Score E
Sequences producing significant alignments: (bits) Value
TC82573 similar to PIR|T09602|T09602 probable zinc finger protei... 306 6e-84
TC77176 homologue to GP|7228329|emb|CAB77055.1 putative TFIIIA (... 249 7e-67
TC86494 homologue to GP|7228329|emb|CAB77055.1 putative TFIIIA (... 159 7e-40
BG648193 similar to GP|2346976|dbj| ZPT2-13 {Petunia x hybrida},... 83 8e-17
AW775690 similar to GP|8567776|gb|A hypothetical protein {Arabid... 83 1e-16
TC81921 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia ... 72 1e-13
TC81612 similar to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia x ... 72 1e-13
TC88653 similar to GP|1786134|dbj|BAA19110.1 PEThy;ZPT2-5 {Petun... 70 9e-13
TC88075 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia ... 67 6e-12
TC93387 similar to GP|1786136|dbj|BAA19111.1 PEThy;ZPT2-6 {Petun... 53 9e-08
AL371585 similar to PIR|S69193|S691 probable finger protein Pszf... 48 3e-06
TC82775 similar to PIR|T52382|T52382 zinc finger protein ZPT4-4 ... 43 1e-04
TC80421 similar to PIR|D96707|D96707 probable zinc finger protei... 42 2e-04
BI308410 similar to PIR|S55882|S558 CCHH finger protein 2 - Arab... 37 0.005
AW686309 homologue to PIR|S55886|S558 CCHH finger protein 6 - Ar... 37 0.007
BI310179 similar to PIR|T52381|T523 zinc finger protein Zat1 C2... 36 0.012
AW690548 similar to SP|Q09134|GRPA Abscisic acid and environment... 35 0.020
TC82737 homologue to PIR|B96686|B96686 probable C2H2-type zinc f... 35 0.020
TC79814 homologue to GP|9759265|dbj|BAB09586.1 gene_id:MRI1.4~pi... 35 0.020
TC89540 similar to PIR|B96686|B96686 probable C2H2-type zinc fin... 35 0.033
>TC82573 similar to PIR|T09602|T09602 probable zinc finger protein SCOF-1
cold-inducible - soybean, partial (65%)
Length = 857
Score = 306 bits (783), Expect = 6e-84
Identities = 166/244 (68%), Positives = 187/244 (76%), Gaps = 8/244 (3%)
Frame = +3
Query: 1 MAMEALNSPTTTAPSFTFNDHPTLRHPAEPWAKRKRSKRSRSCSEEEYLALCLIMLARGG 60
MA+EALNSPT+T P FTF D P L PW KRKRSKRSRSC+EEEYLALCLIMLARG
Sbjct: 39 MALEALNSPTSTTPKFTF-DEPNL-----PWTKRKRSKRSRSCTEEEYLALCLIMLARGH 200
Query: 61 AATTTATPPLQPATA---PSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHS-GGGGED 116
P P T + +++LSYKCSVC+K F SYQALGGHKASHRK+S GGGG+D
Sbjct: 201 TNRHDFNPLNPPPTTIDNNNNNTKLSYKCSVCNKEFSSYQALGGHKASHRKNSVGGGGDD 380
Query: 117 HSAGATSSAVATTTSSANTGSGG-KAHECSICHKSFPTGQALGGHKRCHYEG--GGGSSS 173
H S++ A TTSSANT GG ++HECSICH+SFPTGQALGGHKRCHYEG GGG+S+
Sbjct: 381 HP----STSSAATTSSANTNGGGVRSHECSICHRSFPTGQALGGHKRCHYEGVVGGGASA 548
Query: 174 ATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDEVESPHPV-MKKKQRVFMIPK 232
T SEG+GST HSH RDFDLNIPA PEF + K GEDEVESPHPV MKKK RVF++PK
Sbjct: 549 VTVSEGMGST----HSHQRDFDLNIPAFPEF-ANKVGEDEVESPHPVMMKKKARVFVVPK 713
Query: 233 IEIP 236
IEIP
Sbjct: 714 IEIP 725
>TC77176 homologue to GP|7228329|emb|CAB77055.1 putative TFIIIA (or
kruppel)-like zinc finger protein {Medicago sativa},
partial (96%)
Length = 1005
Score = 249 bits (636), Expect = 7e-67
Identities = 151/247 (61%), Positives = 167/247 (67%), Gaps = 11/247 (4%)
Frame = +2
Query: 1 MAMEALNSPTTTAPSFTFNDHPTLRHPAEPWAKRKRSKRSR----SCSEEEYLALCLIML 56
MAMEALNSPTT P FT + P L + PW K KRSKRSR SC+EEEYLALCLIML
Sbjct: 110 MAMEALNSPTTATP-FTPFEEPNLSYLETPWTKGKRSKRSRMDQSSCTEEEYLALCLIML 286
Query: 57 ARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKH--SGGGG 114
AR G T + P AP + +LS+KCSVC+KAF SYQALGGHKASHRK S
Sbjct: 287 ARSGNNNDNKTESV-PVPAPLTTVKLSHKCSVCNKAFSSYQALGGHKASHRKAVMSATTV 463
Query: 115 EDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEG----GGG 170
ED + TSSAV TTSSA+ G K HECSICHKSFPTGQALGGHKRCHYEG G G
Sbjct: 464 EDQTT-TTSSAV--TTSSASNGKN-KTHECSICHKSFPTGQALGGHKRCHYEGSVGAGAG 631
Query: 171 SSSAT-ASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDEVESPHPVMKKKQRVFM 229
SS+ T ASEGVGS SHSHHRDFDLN+PA P+F +DEV SP P KK
Sbjct: 632 SSAVTAASEGVGS----SHSHHRDFDLNLPAFPDFSKKFFVDDEVSSPLPAAKKP----C 787
Query: 230 IPKIEIP 236
+ K+EIP
Sbjct: 788 LFKLEIP 808
>TC86494 homologue to GP|7228329|emb|CAB77055.1 putative TFIIIA (or
kruppel)-like zinc finger protein {Medicago sativa},
partial (39%)
Length = 1092
Score = 159 bits (403), Expect = 7e-40
Identities = 109/265 (41%), Positives = 141/265 (53%), Gaps = 37/265 (13%)
Frame = +2
Query: 1 MAMEALNSPTTTAPSFTFNDHPTLRHPA----EPWAKRKRSKRSR-----SCSEEEYLAL 51
+ ++ LNS T A + P + E K+KRSKR R +EEEYLAL
Sbjct: 68 LELQTLNSSPTGATTTIPIPFPRFKQEENQERESLVKKKRSKRPRIGIGNPPTEEEYLAL 247
Query: 52 CLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSG 111
CLIML++ ++ PL +L++KCSVC+KAFPSYQALGGHKASHRK S
Sbjct: 248 CLIMLSQSNNQIQSS--PL----------KLNHKCSVCNKAFPSYQALGGHKASHRKSSS 391
Query: 112 GGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEG---- 167
++ + + ++ + S++ K HECSICHKSFPTGQALGGHKRCHYEG
Sbjct: 392 ----ENQSTTVNETISVSVSTS------KMHECSICHKSFPTGQALGGHKRCHYEGVINN 541
Query: 168 -----GGGSSSATASEGVGSTHTVSHSHHRDFDLNIPA-LPEFPS--------------- 206
SS T S+ ++ ++S HR FDLN+PA L EF S
Sbjct: 542 NHNHNNSNSSGITVSDAGAASSSIS---HRGFDLNLPAPLTEFWSPVGFGGGDSKKNKVN 712
Query: 207 ---TKAGEDEVESPHPVMKKKQRVF 228
E EVESP PV K+ R+F
Sbjct: 713 VNLAGINEQEVESPLPVTAKRPRLF 787
>BG648193 similar to GP|2346976|dbj| ZPT2-13 {Petunia x hybrida}, partial
(43%)
Length = 629
Score = 83.2 bits (204), Expect = 8e-17
Identities = 48/116 (41%), Positives = 65/116 (55%)
Frame = +2
Query: 49 LALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRK 108
LA CL+ML+ P Q + AP + Y+C C+K FPS+QALGGH+ASH++
Sbjct: 134 LANCLMMLSYPQHQPQN-NKPNQKSFAP-----VEYECKTCNKKFPSFQALGGHRASHKR 295
Query: 109 HSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCH 164
G E + T ++S + G+ K HECSIC ++F GQALGGH R H
Sbjct: 296 SKLEGDE----------LLTNSTSLSLGNKPKMHECSICGQNFSLGQALGGHMRRH 433
Score = 30.8 bits (68), Expect = 0.48
Identities = 12/30 (40%), Positives = 18/30 (60%)
Frame = +2
Query: 78 GSSRLSYKCSVCDKAFPSYQALGGHKASHR 107
G+ ++CS+C + F QALGGH H+
Sbjct: 347 GNKPKMHECSICGQNFSLGQALGGHMRRHK 436
>AW775690 similar to GP|8567776|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (20%)
Length = 631
Score = 82.8 bits (203), Expect = 1e-16
Identities = 56/154 (36%), Positives = 70/154 (45%), Gaps = 25/154 (16%)
Frame = +2
Query: 36 RSKRSRSCSEEEYLALCLIMLARGGAAT----TTATPPLQPATAPSGSSRLSYKCSVCDK 91
R + EEE LA CLI+LARG T + S+ Y+C C++
Sbjct: 140 RDRSEEEKEEEEDLANCLILLARGHNHRHDYQNTHVSNHRDHNNNKSSNFYLYECKTCNR 319
Query: 92 AFPSYQALGGHKASHRKHSGGG---GEDHSAGATSSAV------------------ATTT 130
FPS+QALGGH+ASH K + + A TSS V AT
Sbjct: 320 CFPSFQALGGHRASHTKPNKANCNVAQQKQAVTTSSFVDDHYDPTMNTILSLQAFAATPI 499
Query: 131 SSANTGSGGKAHECSICHKSFPTGQALGGHKRCH 164
++ T K HECSIC F +GQALGGH R H
Sbjct: 500 TTIPTTKKSKVHECSICGAEFSSGQALGGHMRRH 601
Score = 35.8 bits (81), Expect = 0.015
Identities = 22/76 (28%), Positives = 36/76 (46%)
Frame = +2
Query: 32 AKRKRSKRSRSCSEEEYLALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDK 91
A++K++ + S ++ Y +L+ A T T T P+ ++CS+C
Sbjct: 395 AQQKQAVTTSSFVDDHYDPTMNTILSLQAFAATPIT------TIPTTKKSKVHECSICGA 556
Query: 92 AFPSYQALGGHKASHR 107
F S QALGGH H+
Sbjct: 557 EFSSGQALGGHMRRHK 604
>TC81921 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia x hybrida},
partial (41%)
Length = 839
Score = 72.4 bits (176), Expect = 1e-13
Identities = 48/150 (32%), Positives = 67/150 (44%), Gaps = 20/150 (13%)
Frame = +3
Query: 49 LALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRK 108
+A CL++L+RG + + + + S+ ++C C++ FPS+QALGGH+ASH+K
Sbjct: 159 MANCLMLLSRG-------SDQFEATYSSTTSNNRVFECKTCNRQFPSFQALGGHRASHKK 317
Query: 109 HSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCH---- 164
G +T K HECSIC F GQALGGH R H
Sbjct: 318 PRLMGEN------------IDGQLLHTPPKPKTHECSICGLEFAIGQALGGHMRRHRAAN 461
Query: 165 ----------------YEGGGGSSSATASE 178
GGGG SS +S+
Sbjct: 462 MNGNKNMHNSNNTMSCSSGGGGDSSIDSSQ 551
>TC81612 similar to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia x hybrida},
partial (50%)
Length = 612
Score = 72.4 bits (176), Expect = 1e-13
Identities = 40/103 (38%), Positives = 53/103 (50%)
Frame = +3
Query: 84 YKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHE 143
Y+C C+K F S+QALGGH+ASH++ GE+ + S + + K HE
Sbjct: 192 YECKTCNKKFSSFQALGGHRASHKRMKLAEGEE---------LKEQAKSLSLWNKPKMHE 344
Query: 144 CSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTV 186
CSIC F GQALGGH R H A +EGV S + +
Sbjct: 345 CSICGMGFSLGQALGGHMRKH--------RAVINEGVSSINQI 449
>TC88653 similar to GP|1786134|dbj|BAA19110.1 PEThy;ZPT2-5 {Petunia x
hybrida}, partial (59%)
Length = 866
Score = 69.7 bits (169), Expect = 9e-13
Identities = 40/105 (38%), Positives = 55/105 (52%), Gaps = 4/105 (3%)
Frame = +1
Query: 84 YKCSVCDKAFPSYQALGGHKASHRK----HSGGGGEDHSAGATSSAVATTTSSANTGSGG 139
++C C + F S+QALGGH+ASH+K G+DH ++ T+T+ A
Sbjct: 151 FECKTCKRQFSSFQALGGHRASHKKPRLMEMTSDGDDH-----HGSILTSTTKA------ 297
Query: 140 KAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTH 184
K H CSIC F GQALGGH R H + S+ A+ G+ H
Sbjct: 298 KTHACSICGLEFGIGQALGGHMRRHRR----TESSKANNSNGNMH 420
Score = 41.6 bits (96), Expect = 3e-04
Identities = 19/54 (35%), Positives = 28/54 (51%)
Frame = +1
Query: 83 SYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTG 136
++ CS+C F QALGGH HR+ + + + TTTSS+N+G
Sbjct: 301 THACSICGLEFGIGQALGGHMRRHRRTESSKANNSNGNMHNFMTTTTTSSSNSG 462
Score = 30.8 bits (68), Expect = 0.48
Identities = 11/25 (44%), Positives = 15/25 (60%)
Frame = +1
Query: 140 KAHECSICHKSFPTGQALGGHKRCH 164
+ EC C + F + QALGGH+ H
Sbjct: 145 RVFECKTCKRQFSSFQALGGHRASH 219
>TC88075 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia x hybrida},
partial (38%)
Length = 674
Score = 67.0 bits (162), Expect = 6e-12
Identities = 53/186 (28%), Positives = 82/186 (43%), Gaps = 8/186 (4%)
Frame = +3
Query: 24 LRHPAEPWAKRKRSKRSRSCSEEEYLALCLIMLARGGAATTTATPPLQPATAPSGSSRLS 83
L H P KR+R + + LA L++L+ P++
Sbjct: 9 LSHTLIPMMKRQRENEEKESID---LATTLMLLSSRITQQNKTYSPVE------------ 143
Query: 84 YKCSVCDKAFPSYQALGGHKASHRKHSGGGGED-HSAGATSSAVATTTSSANTGSGGKAH 142
++C C++ F S+QALGGH+ASH+K E+ H ++ K H
Sbjct: 144 FECKTCNRKFSSFQALGGHRASHKKLKLLDSEEAHKVNIHNNKP-------------KMH 284
Query: 143 ECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGV--GSTHTVSHSHHR-----DFD 195
+CSIC + F GQALGGH R H G SS++ + V S+ + S+ + + D
Sbjct: 285 QCSICGQEFKLGQALGGHMRRHRINNEGFSSSSINYQVIAKSSPVLKRSNSKRVMIMELD 464
Query: 196 LNIPAL 201
LN+ L
Sbjct: 465 LNLTPL 482
>TC93387 similar to GP|1786136|dbj|BAA19111.1 PEThy;ZPT2-6 {Petunia x
hybrida}, partial (17%)
Length = 650
Score = 53.1 bits (126), Expect = 9e-08
Identities = 29/76 (38%), Positives = 41/76 (53%), Gaps = 12/76 (15%)
Frame = +1
Query: 45 EEEYLALCLIMLARGGAATTTA--TPPLQPATAPSGSSRLS----------YKCSVCDKA 92
EEE LA CLI+LA+GG + G+ ++ Y+C C++
Sbjct: 319 EEEDLAKCLILLAQGGNHREDGGVVDENKRVKGSHGNKKIGETSTKLGLYIYECKTCNRT 498
Query: 93 FPSYQALGGHKASHRK 108
FPS+QALGGH+ASH+K
Sbjct: 499 FPSFQALGGHRASHKK 546
Score = 40.0 bits (92), Expect = 8e-04
Identities = 36/133 (27%), Positives = 57/133 (42%)
Frame = +1
Query: 32 AKRKRSKRSRSCSEEEYLALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDK 91
AK KR+KR R +L+ C ++ ++ + L+ L+ KC +
Sbjct: 199 AKGKRTKRLR------FLSPCGVVAMASSCSSDESNIDLEDEDEDDEEEDLA-KCLIL-- 351
Query: 92 AFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSF 151
A GG+ HR+ GG D + S +T G +EC C+++F
Sbjct: 352 -----LAQGGN---HRED--GGVVDENKRVKGSHGNKKIGETSTKLGLYIYECKTCNRTF 501
Query: 152 PTGQALGGHKRCH 164
P+ QALGGH+ H
Sbjct: 502 PSFQALGGHRASH 540
>AL371585 similar to PIR|S69193|S691 probable finger protein Pszf1 - garden
pea, partial (19%)
Length = 497
Score = 48.1 bits (113), Expect = 3e-06
Identities = 32/85 (37%), Positives = 40/85 (46%), Gaps = 8/85 (9%)
Frame = +1
Query: 132 SANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTVSHSHH 191
+A G+ K HECSIC F +GQALGGH R H S A A+ V +
Sbjct: 40 AAINGNKAKIHECSICGSEFTSGQALGGHMRRH-----RVSVANAAAVAAPDERVRPRNI 204
Query: 192 RDFDLNIPALPE--------FPSTK 208
DLN+PA E FP+T+
Sbjct: 205 LQLDLNLPAPEEDIRESKFQFPATQ 279
Score = 35.0 bits (79), Expect = 0.026
Identities = 15/33 (45%), Positives = 20/33 (60%)
Frame = +1
Query: 75 APSGSSRLSYKCSVCDKAFPSYQALGGHKASHR 107
A +G+ ++CS+C F S QALGGH HR
Sbjct: 43 AINGNKAKIHECSICGSEFTSGQALGGHMRRHR 141
>TC82775 similar to PIR|T52382|T52382 zinc finger protein ZPT4-4 C2H2-type
[imported] - garden petunia, partial (17%)
Length = 1152
Score = 42.7 bits (99), Expect = 1e-04
Identities = 29/109 (26%), Positives = 41/109 (37%), Gaps = 26/109 (23%)
Frame = +3
Query: 84 YKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAG----ATSSAVATTTSSANTGSGG 139
+ C C K+FP ++LGGH SH + +H +T + + +A GS G
Sbjct: 150 HACKFCSKSFPCGRSLGGHMRSHINNLSAEKAEHKEKQLIFSTKKNDSIGSEAATNGSYG 329
Query: 140 KAHE----------------------CSICHKSFPTGQALGGHKRCHYE 166
C C K F + QAL GH +CH E
Sbjct: 330 LRENPKKTWRKRIADSSEDSYVYDKFCKECGKCFHSWQALFGHMKCHSE 476
>TC80421 similar to PIR|D96707|D96707 probable zinc finger protein T22E19.1
[imported] - Arabidopsis thaliana, partial (37%)
Length = 1217
Score = 42.0 bits (97), Expect = 2e-04
Identities = 21/54 (38%), Positives = 27/54 (49%), Gaps = 1/54 (1%)
Frame = +2
Query: 112 GGGEDHSAGATSSAVATT-TSSANTGSGGKAHECSICHKSFPTGQALGGHKRCH 164
G G +G+T S SS N SG + +EC C + F QALGGH+ H
Sbjct: 155 GTGNTSPSGSTESVSGDGGASSINPTSGDRKYECQYCCREFANSQALGGHQNAH 316
Score = 35.0 bits (79), Expect = 0.026
Identities = 16/56 (28%), Positives = 29/56 (51%), Gaps = 7/56 (12%)
Frame = +2
Query: 60 GAATTTATPPLQPATAPSGSSRLS-------YKCSVCDKAFPSYQALGGHKASHRK 108
G T+ + + + G+S ++ Y+C C + F + QALGGH+ +H+K
Sbjct: 155 GTGNTSPSGSTESVSGDGGASSINPTSGDRKYECQYCCREFANSQALGGHQNAHKK 322
>BI308410 similar to PIR|S55882|S558 CCHH finger protein 2 - Arabidopsis
thaliana, partial (46%)
Length = 492
Score = 37.4 bits (85), Expect = 0.005
Identities = 38/143 (26%), Positives = 61/143 (42%), Gaps = 14/143 (9%)
Frame = +3
Query: 70 LQPATAPSGSS----RLSYKCSVCDKAFPSYQALGGHKASH---RKHSGGGGEDHSAGAT 122
L+P+++ S SS + + C+ C + F S QALGGH+ +H R + E SA +
Sbjct: 48 LEPSSSSSSSSSSMEQRIFSCNYCQRKFYSSQALGGHQNAHKLERTLAKKSREMSSAMQS 227
Query: 123 SSA-----VATTTSSANTGSGGKAHECSICHKSFPTGQAL--GGHKRCHYEGGGGSSSAT 175
S A + +++ + GS G AH ++ G + GG K G A+
Sbjct: 228 SYAELPEHPSNFSTNYHLGSHGNAH----LDNNYRQGHVMRHGGRKDQFSYGNSKEGGAS 395
Query: 176 ASEGVGSTHTVSHSHHRDFDLNI 198
S G S DL++
Sbjct: 396 WSRGYNSNSENVQEDISQLDLSL 464
>AW686309 homologue to PIR|S55886|S558 CCHH finger protein 6 - Arabidopsis
thaliana, partial (25%)
Length = 669
Score = 37.0 bits (84), Expect = 0.007
Identities = 15/37 (40%), Positives = 21/37 (56%)
Frame = +2
Query: 128 TTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCH 164
T + S+ +G K +EC C + F QALGGH+ H
Sbjct: 185 TQSESSEPLNGRKLYECQYCCREFANSQALGGHQNAH 295
Score = 36.2 bits (82), Expect = 0.012
Identities = 15/40 (37%), Positives = 24/40 (59%)
Frame = +2
Query: 69 PLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRK 108
P Q ++ + R Y+C C + F + QALGGH+ +H+K
Sbjct: 182 PTQSESSEPLNGRKLYECQYCCREFANSQALGGHQNAHKK 301
>BI310179 similar to PIR|T52381|T523 zinc finger protein Zat1 C2H2-type
[imported] - Arabidopsis thaliana, partial (16%)
Length = 704
Score = 36.2 bits (82), Expect = 0.012
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = +2
Query: 142 HECSICHKSFPTGQALGGHKRCH 164
H+C C +SF G+ALGGH R H
Sbjct: 77 HKCKFCMRSFSNGRALGGHMRSH 145
Score = 30.0 bits (66), Expect = 0.82
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = +2
Query: 84 YKCSVCDKAFPSYQALGGHKASH 106
+KC C ++F + +ALGGH SH
Sbjct: 77 HKCKFCMRSFSNGRALGGHMRSH 145
>AW690548 similar to SP|Q09134|GRPA Abscisic acid and environmental stress
inducible protein. [Sickle medic] {Medicago falcata},
partial (68%)
Length = 599
Score = 35.4 bits (80), Expect = 0.020
Identities = 34/121 (28%), Positives = 39/121 (32%), Gaps = 21/121 (17%)
Frame = +1
Query: 95 SYQALGGHKASHRKHSGGGGEDHSAG-------------ATSSAVAT-----TTSSANTG 136
+Y GGH H GGGG H G A AV T + A G
Sbjct: 187 NYHNGGGHYYGGGSHHGGGGSHHGGGGCHYYCHGHCCSYAEFVAVQTEEKTNEVNDAKYG 366
Query: 137 SGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTVSHSH---HRD 193
GG + H GG HY GGG + G G H H H H +
Sbjct: 367 GGGYHNGGGNYHNG-------GG----HYYGGGSHGGGGSHHGGGGCHYYCHGHCCSHAE 513
Query: 194 F 194
F
Sbjct: 514 F 516
>TC82737 homologue to PIR|B96686|B96686 probable C2H2-type zinc finger
protein F15E12.19 [imported] - Arabidopsis thaliana,
partial (18%)
Length = 1006
Score = 35.4 bits (80), Expect = 0.020
Identities = 14/44 (31%), Positives = 26/44 (58%)
Frame = +2
Query: 121 ATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCH 164
+T + ++T+ S+N + + C+ C + F + QALGGH+ H
Sbjct: 350 STGISFSSTSESSNELTIQRVFSCNYCRRKFYSSQALGGHQNAH 481
Score = 33.1 bits (74), Expect = 0.097
Identities = 22/90 (24%), Positives = 41/90 (45%), Gaps = 4/90 (4%)
Frame = +2
Query: 23 TLRHPAEPWAKRKRSKRSRSCSEEEYLALCLIMLARGGAATTTATPPLQPATAPSGSSRL 82
T+ HP P + S S S + ++L L + + + ++ S+ L
Sbjct: 224 TISHP--PN*NLYKGSYSNSLSNSDSISLDLTLNFNNNDLVVRDSTGISFSSTSESSNEL 397
Query: 83 S----YKCSVCDKAFPSYQALGGHKASHRK 108
+ + C+ C + F S QALGGH+ +H++
Sbjct: 398 TIQRVFSCNYCRRKFYSSQALGGHQNAHKR 487
>TC79814 homologue to GP|9759265|dbj|BAB09586.1
gene_id:MRI1.4~pir||T04230~similar to unknown protein
{Arabidopsis thaliana}, partial (42%)
Length = 892
Score = 35.4 bits (80), Expect = 0.020
Identities = 28/101 (27%), Positives = 46/101 (44%), Gaps = 6/101 (5%)
Frame = +3
Query: 38 KRSRSCSEEEYLALCLIMLARGGAATTTATPPLQPATAPSGSSRLSY--KCSVC----DK 91
+R+ ++ Y L+++A T ATP + +T+ S S S+ S C DK
Sbjct: 345 ERNNDTDQKPYEEKILVIMAGQDKPTFLATPSMSSSTSTSTSRSSSFGDNTSTCTCEHDK 524
Query: 92 AFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSS 132
S + + +++ K GGGGE+H S TT +S
Sbjct: 525 DQKSIENMNDEESTV-KQGGGGGENHVRRTESVETPTTDTS 644
>TC89540 similar to PIR|B96686|B96686 probable C2H2-type zinc finger protein
F15E12.19 [imported] - Arabidopsis thaliana, partial
(28%)
Length = 1639
Score = 34.7 bits (78), Expect = 0.033
Identities = 17/49 (34%), Positives = 26/49 (52%)
Frame = +1
Query: 60 GAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRK 108
GAA A+P + P + C+ C + F S QALGGH+ +H++
Sbjct: 532 GAAEVHASPSVTPRI---------FSCNYCKRKFYSSQALGGHQNAHKR 651
Score = 31.6 bits (70), Expect = 0.28
Identities = 14/42 (33%), Positives = 20/42 (47%)
Frame = +1
Query: 123 SSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCH 164
SS V A+ + C+ C + F + QALGGH+ H
Sbjct: 520 SSEVGAAEVHASPSVTPRIFSCNYCKRKFYSSQALGGHQNAH 645
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.126 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,316,003
Number of Sequences: 36976
Number of extensions: 126586
Number of successful extensions: 1032
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 927
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 982
length of query: 239
length of database: 9,014,727
effective HSP length: 93
effective length of query: 146
effective length of database: 5,575,959
effective search space: 814090014
effective search space used: 814090014
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0096a.2


