

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
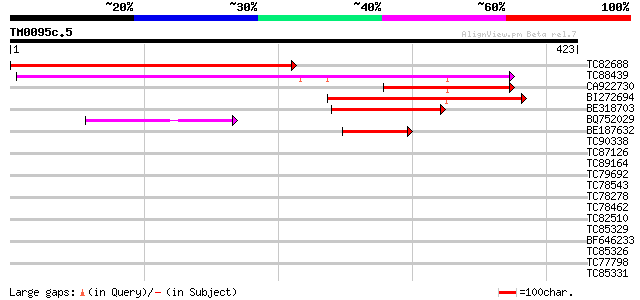
Query= TM0095c.5
(423 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82688 weakly similar to GP|13477193|gb|AAH05058.1 Similar to C... 368 e-102
TC88439 weakly similar to PIR|E84546|E84546 probable tetracyclin... 246 1e-65
CA922730 similar to PIR|F84546|F845 probable tetracycline transp... 162 2e-40
BI272694 weakly similar to PIR|G84546|G845 probable tetracycline... 124 7e-29
BE318703 weakly similar to GP|10177006|dbj contains similarity t... 88 7e-18
BQ752029 GP|12002184|g tetracycline resistance protein {IncN pla... 49 4e-06
BE187632 weakly similar to PIR|F86258|F862 protein F5O11.14 [imp... 49 5e-06
TC90338 similar to GP|10178069|dbj|BAB11433. emb|CAB89350.1~gene... 32 0.46
TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {... 30 1.8
TC89164 weakly similar to GP|10177340|dbj|BAB10596. transporter-... 29 3.0
TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 29 3.9
TC78543 similar to PIR|T14545|T14545 probable sugar transporter ... 29 3.9
TC78278 homologue to GP|12321869|gb|AAG50965.1 integral membrane... 28 5.1
TC78462 similar to GP|21592780|gb|AAM64729.1 nucleic acid bindin... 28 5.1
TC82510 weakly similar to GP|14532868|gb|AAK64116.1 unknown prot... 28 6.7
TC85329 homologue to GP|5738542|emb|CAB53034.1 photosystem I sub... 28 6.7
BF646233 similar to GP|14165163|gb| 2-on-2 hemoglobin {Arabidops... 28 8.7
TC85326 similar to GP|5738542|emb|CAB53034.1 photosystem I subun... 28 8.7
TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 28 8.7
TC85331 homologue to GP|5738542|emb|CAB53034.1 photosystem I sub... 28 8.7
>TC82688 weakly similar to GP|13477193|gb|AAH05058.1 Similar to CG5078 gene
product {Homo sapiens}, partial (30%)
Length = 734
Score = 368 bits (945), Expect = e-102
Identities = 181/214 (84%), Positives = 195/214 (90%)
Frame = +3
Query: 1 MSWWSGLIELRPLFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPGGSTCPKVIYINGLQQ 60
MSWWSG ELRPLFHLL PLSIHWIAEEMTVSVLVDVTT+ALCP S+C K IYINGLQ+
Sbjct: 75 MSWWSGFFELRPLFHLLLPLSIHWIAEEMTVSVLVDVTTTALCPQQSSCSKAIYINGLQE 254
Query: 61 TIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFS 120
TI GIFKM+VLPLLGQLSD+HGRKP LL+TMSTTIF FA+LAW+QSEEFVYAYYVLRT S
Sbjct: 255 TIAGIFKMMVLPLLGQLSDDHGRKPFLLLTMSTTIFPFALLAWNQSEEFVYAYYVLRTIS 434
Query: 121 YIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSI 180
YIIS+GSIFCISVAYVA+VVNE+KRAAVF WITGLFSASHVVGNVLARFLP+ YIFVVSI
Sbjct: 435 YIISKGSIFCISVAYVANVVNENKRAAVFGWITGLFSASHVVGNVLARFLPQNYIFVVSI 614
Query: 181 ALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKV 214
ALL FCPVYMQFFLVET AP+KNQ GFC+KV
Sbjct: 615 ALLIFCPVYMQFFLVETGKLAPRKNQELGFCSKV 716
>TC88439 weakly similar to PIR|E84546|E84546 probable tetracycline
transporter protein [imported] - Arabidopsis thaliana,
partial (37%)
Length = 1399
Score = 246 bits (628), Expect = 1e-65
Identities = 138/411 (33%), Positives = 225/411 (54%), Gaps = 40/411 (9%)
Frame = +2
Query: 6 GLIELRPLFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPGGSTCPKVIYINGLQQTIVGI 65
G+ L L HL + + + + + DVT +ALCPG C IY++G QQ ++GI
Sbjct: 32 GMEGLTGLSHLFVTMFLAGFGGIIVMPSITDVTMAALCPGQDECSLAIYLSGFQQVMIGI 211
Query: 66 FKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQ 125
++ PL+G LSD +GRK L+ + ++ ++ A+LA+ + +F YAYYV++T + + +
Sbjct: 212 GSVMTTPLIGNLSDRYGRKALITLPLTVSLIPQAILAYSRDTKFFYAYYVVKTLAALAGE 391
Query: 126 GSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTF 185
GS C+++AYVAD+V KRA+ F + G+ SAS V G + ARFL F V+
Sbjct: 392 GSFHCLALAYVADIVPVGKRASAFGILAGVGSASFVGGTLAARFLSAALTFQVASVSSMI 571
Query: 186 CPVYMQFFLVETV-TPAPKKNQVSGFCTKVV--DVLHKRYKSMRNAAEILDNL------- 235
VYM+ FL E+ P + C + D+ + +K + + +++ L
Sbjct: 572 GLVYMRIFLKESAPMRQPLLKEAEEPCIEQCEDDLPQRTFKKLPSMGDLICLLKCSPTFS 751
Query: 236 ----------------IIAVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPK 279
+ YYLKA F +NKNQF++L+M+ GIG+ +Q+ L+PIL P
Sbjct: 752 QAAIVLLFNSLADGGSMAVTMYYLKARFQYNKNQFADLMMISGIGATLTQLFLMPILVPA 931
Query: 280 VGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKPS-------------- 325
VGE+ +L+ L S +Y ++W+ WVPY A F ++ VLV+PS
Sbjct: 932 VGEEKLLTTGLFVSCISMLMYSISWSAWVPYALAGFSVLGVLVRPSLTSIASKQVGPNEQ 1111
Query: 326 GKAQTFIAGAQSISDLLSPIAMSPLTSWFLSSNAPFDCKGFSIICASVSMV 376
G Q ++G S ++++SP+ SPLT+ FLS +APF+ GFS++C ++++
Sbjct: 1112GMVQGCLSGITSAANIISPLIFSPLTALFLSEDAPFNFPGFSLMCLGLTLM 1264
>CA922730 similar to PIR|F84546|F845 probable tetracycline transporter
protein [imported] - Arabidopsis thaliana, partial (6%)
Length = 757
Score = 162 bits (410), Expect = 2e-40
Identities = 85/111 (76%), Positives = 91/111 (81%), Gaps = 14/111 (12%)
Frame = -2
Query: 280 VGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKPS-------------- 325
VGEKVIL +ALLASIAYAWL GLAWAPWVPY SASFGIIYVLVKP+
Sbjct: 756 VGEKVILCSALLASIAYAWLSGLAWAPWVPYLSASFGIIYVLVKPATYAIISRASSSTNQ 577
Query: 326 GKAQTFIAGAQSISDLLSPIAMSPLTSWFLSSNAPFDCKGFSIICASVSMV 376
GKAQTFIAGAQSISDLLSPI MSPLTS FLSS+APF+CKGFSI+CASV M+
Sbjct: 576 GKAQTFIAGAQSISDLLSPIVMSPLTSLFLSSDAPFECKGFSILCASVCMM 424
>BI272694 weakly similar to PIR|G84546|G845 probable tetracycline transporter
protein [imported] - Arabidopsis thaliana, partial (17%)
Length = 648
Score = 124 bits (311), Expect = 7e-29
Identities = 66/163 (40%), Positives = 99/163 (60%), Gaps = 15/163 (9%)
Frame = +1
Query: 238 AVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYA 297
++ YYLKA F F+KN F++L+++ GI SQ+ L+PI P +GE +LS L +
Sbjct: 61 SMMYYLKARFHFDKNHFADLMIISGIAGTVSQLFLMPIFAPTLGEARLLSIGLFFHCVHM 240
Query: 298 WLYGLAWAPWVPYFSASFGIIYVLVKP--------------SGKAQTFIAGAQSISDLLS 343
++Y +AW+ WVPY +A F I++V +P G+AQ I+G SI+ ++S
Sbjct: 241 FIYSIAWSSWVPYAAAMFSILFVFSQPCIRSIVSKQVDPREQGRAQGCISGICSIAHIVS 420
Query: 344 PIAMSPLTSWFLSSNAPFDCKGFSIICASV-SMVAETENEKLK 385
P+A SPLT+ FLS APF+ GFSI+C + SMV+ + LK
Sbjct: 421 PLAFSPLTALFLSEKAPFNFPGFSIMCIGIASMVSFVXSMMLK 549
>BE318703 weakly similar to GP|10177006|dbj contains similarity to
tetracycline transporter protein~gene_id:K5J14.1
{Arabidopsis thaliana}, partial (17%)
Length = 588
Score = 87.8 bits (216), Expect = 7e-18
Identities = 39/85 (45%), Positives = 58/85 (67%)
Frame = -2
Query: 241 YYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLY 300
YYLKA F +NQF++L+M+ GIG+ +Q+ L+PIL P VGE+ +L+ L S +Y
Sbjct: 542 YYLKAPFSIQQNQFADLMMISGIGATLTQLFLMPILVPAVGEEKLLTTGLFVSCISMLMY 363
Query: 301 GLAWAPWVPYFSASFGIIYVLVKPS 325
++W+ WVPY A F ++ VLV+PS
Sbjct: 362 SISWSAWVPYALAGFSVLGVLVRPS 288
>BQ752029 GP|12002184|g tetracycline resistance protein {IncN plasmid R46},
partial (47%)
Length = 645
Score = 48.9 bits (115), Expect = 4e-06
Identities = 29/114 (25%), Positives = 54/114 (46%)
Frame = +2
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVL 116
G+ + + + + P+LG LSD GR+P+LL ++ +A++A +YA ++
Sbjct: 218 GVLLALYALMQFLCAPVLGALSDRFGRRPVLLASLLGATIDYAIMATTPVLWILYAGRIV 397
Query: 117 RTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFL 170
+ G+ ++ AY+AD+ + RA F ++ F V G V L
Sbjct: 398 AGIT-----GATGAVAGAYIADITDGEDRARHFGLMSACFGVGMVAGPVAGGLL 544
>BE187632 weakly similar to PIR|F86258|F862 protein F5O11.14 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 606
Score = 48.5 bits (114), Expect = 5e-06
Identities = 28/52 (53%), Positives = 34/52 (64%)
Frame = +3
Query: 249 FNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLY 300
F + FS L + S +S +V+LPILNP VGEKVIL +ALLASI LY
Sbjct: 216 FFRCNFSNSLFPSCLNSRYSPIVMLPILNPLVGEKVILFSALLASILLGVLY 371
>TC90338 similar to GP|10178069|dbj|BAB11433.
emb|CAB89350.1~gene_id:MUB3.18~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (18%)
Length = 734
Score = 32.0 bits (71), Expect = 0.46
Identities = 17/39 (43%), Positives = 20/39 (50%)
Frame = +1
Query: 6 GLIELRPLFHLLFPLSIHWIAEEMTVSVLVDVTTSALCP 44
G I L LFH LFPLS + E V + + V T CP
Sbjct: 1 GSISLFVLFHSLFPLSSTTMVRERVVDLCITVPTFFKCP 117
>TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {Arabidopsis
thaliana}, partial (79%)
Length = 1524
Score = 30.0 bits (66), Expect = 1.8
Identities = 18/76 (23%), Positives = 37/76 (48%)
Frame = +3
Query: 75 GQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVA 134
GQ+++ GRK L+I I + +++ + F++ +L F G I +
Sbjct: 33 GQIAEYVGRKGSLMIASIPNIIGWLAISFAKDSSFLFMGRLLEGFGV----GIISYVVPV 200
Query: 135 YVADVVNESKRAAVFS 150
Y+A++ E+ R ++ S
Sbjct: 201 YIAEIAPENMRGSLGS 248
>TC89164 weakly similar to GP|10177340|dbj|BAB10596. transporter-like
protein {Arabidopsis thaliana}, partial (49%)
Length = 1129
Score = 29.3 bits (64), Expect = 3.0
Identities = 19/105 (18%), Positives = 47/105 (44%), Gaps = 5/105 (4%)
Frame = +3
Query: 73 LLGQLSDEHGRKPLLL-----ITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGS 127
L G +SD +GRKP+++ + + T+F + W + ++L + + ++
Sbjct: 537 LWGMVSDRYGRKPVIIMGIIAVVIFNTLFGLSTGYW----MAIITRFLLGSLNGVLGPVK 704
Query: 128 IFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPE 172
AY +++ E +A S ++ + ++G + +L +
Sbjct: 705 ------AYASELFREEHQAIGLSTVSAAWGVGLIIGPAIGGYLAQ 821
>TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (49%)
Length = 1353
Score = 28.9 bits (63), Expect = 3.9
Identities = 17/49 (34%), Positives = 24/49 (48%)
Frame = +2
Query: 55 INGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAW 103
+ GL + I VV G LSD GR+P+L+I+ S V+ W
Sbjct: 281 VEGLIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFW 427
>TC78543 similar to PIR|T14545|T14545 probable sugar transporter protein -
beet, partial (93%)
Length = 1653
Score = 28.9 bits (63), Expect = 3.9
Identities = 18/76 (23%), Positives = 36/76 (46%)
Frame = +1
Query: 75 GQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVA 134
GQ+++ GRK L+I I + ++++ F+Y +L F G I
Sbjct: 322 GQIAEYMGRKGSLMIASIPNIIGWLMISFANDSSFLYMGRLLEGFGV----GIISYTVPV 489
Query: 135 YVADVVNESKRAAVFS 150
Y+A++ ++ R ++ S
Sbjct: 490 YIAEISPQNLRGSLVS 537
>TC78278 homologue to GP|12321869|gb|AAG50965.1 integral membrane protein
putative; 85705-84183 {Arabidopsis thaliana}, partial
(87%)
Length = 1415
Score = 28.5 bits (62), Expect = 5.1
Identities = 28/126 (22%), Positives = 54/126 (42%), Gaps = 17/126 (13%)
Frame = +2
Query: 66 FKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFA-VLAWDQSE------------EFVYA 112
F + ++ LL D+ G K + ST F+ V AW S + +
Sbjct: 257 FLITIMKLLQVSEDQKGSKTMK--GASTRFFTIGLVAAWYSSNIGVLLLNKYLLSNYGFK 430
Query: 113 YYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAV----FSWITGLFSASHVVGNVLAR 168
Y + T ++ + +++A++ V ++ R+ V S ++ +F S V GN+ R
Sbjct: 431 YPIFLTMCHMTACSLFSYVAIAWMKIVPMQTIRSRVQFFKISALSLIFCVSVVFGNISLR 610
Query: 169 FLPEEY 174
+LP +
Sbjct: 611 YLPVSF 628
>TC78462 similar to GP|21592780|gb|AAM64729.1 nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (85%)
Length = 1281
Score = 28.5 bits (62), Expect = 5.1
Identities = 12/29 (41%), Positives = 18/29 (61%)
Frame = +3
Query: 233 DNLIIAVQYYLKAVFGFNKNQFSELLMMV 261
D+ +IA+ YY A FGF+K+ L M+
Sbjct: 498 DSWLIAIAYYFGARFGFDKSDRKRLFNMI 584
>TC82510 weakly similar to GP|14532868|gb|AAK64116.1 unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 1716
Score = 28.1 bits (61), Expect = 6.7
Identities = 13/31 (41%), Positives = 22/31 (70%)
Frame = +1
Query: 378 ETENEKLKENLQDEKNKNIELEKQMIKLKAQ 408
+ +NEK+KE+LQ + + +ELEK K++ Q
Sbjct: 1483 QPDNEKMKEHLQGMQWRVMELEKVCKKMQTQ 1575
>TC85329 homologue to GP|5738542|emb|CAB53034.1 photosystem I subunit XI
precursor {Arabidopsis thaliana}, partial (79%)
Length = 1068
Score = 28.1 bits (61), Expect = 6.7
Identities = 13/37 (35%), Positives = 22/37 (59%)
Frame = +1
Query: 321 LVKPSGKAQTFIAGAQSISDLLSPIAMSPLTSWFLSS 357
L++P+ + I G I L +P+ SPL +W+LS+
Sbjct: 460 LLQPNYQVIQPINGDPFIGSLETPVTSSPLVAWYLSN 570
>BF646233 similar to GP|14165163|gb| 2-on-2 hemoglobin {Arabidopsis
thaliana}, partial (40%)
Length = 581
Score = 27.7 bits (60), Expect = 8.7
Identities = 14/50 (28%), Positives = 27/50 (54%), Gaps = 7/50 (14%)
Frame = -1
Query: 80 EHGRKPLLLITMSTTIFSFAVLAWDQSEE-------FVYAYYVLRTFSYI 122
E+ KPLL++ +++ I+S + + SEE ++Y Y + + YI
Sbjct: 317 EY*TKPLLIVNINSIIYSTTINYHEHSEERLFNIEIYIYIYIYIYIYIYI 168
>TC85326 similar to GP|5738542|emb|CAB53034.1 photosystem I subunit XI
precursor {Arabidopsis thaliana}, partial (85%)
Length = 826
Score = 27.7 bits (60), Expect = 8.7
Identities = 13/35 (37%), Positives = 20/35 (57%)
Frame = +1
Query: 323 KPSGKAQTFIAGAQSISDLLSPIAMSPLTSWFLSS 357
KP+ + I G I L +P+ SPL +W+LS+
Sbjct: 196 KPNYQVIQPINGDPFIGSLETPVTSSPLVAWYLSN 300
>TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (88%)
Length = 2467
Score = 27.7 bits (60), Expect = 8.7
Identities = 15/43 (34%), Positives = 25/43 (57%), Gaps = 3/43 (6%)
Frame = +3
Query: 62 IVGIFKMVVLPLLG---QLSDEHGRKPLLLITMSTTIFSFAVL 101
I + +++LP +G +L D GR+ LLL+T+ I S +L
Sbjct: 1782 ISAVTTLLMLPSIGLAMRLMDVTGRRQLLLVTIPVLIVSLVIL 1910
>TC85331 homologue to GP|5738542|emb|CAB53034.1 photosystem I subunit XI
precursor {Arabidopsis thaliana}, partial (75%)
Length = 662
Score = 27.7 bits (60), Expect = 8.7
Identities = 13/35 (37%), Positives = 20/35 (57%)
Frame = +2
Query: 323 KPSGKAQTFIAGAQSISDLLSPIAMSPLTSWFLSS 357
KP+ + I G I L +P+ SPL +W+LS+
Sbjct: 158 KPNYQGIQPINGDPYIGSLETPVTSSPLVAWYLSN 262
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,068,002
Number of Sequences: 36976
Number of extensions: 197976
Number of successful extensions: 1432
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1420
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1427
length of query: 423
length of database: 9,014,727
effective HSP length: 99
effective length of query: 324
effective length of database: 5,354,103
effective search space: 1734729372
effective search space used: 1734729372
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0095c.5


