

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093b.6
(443 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
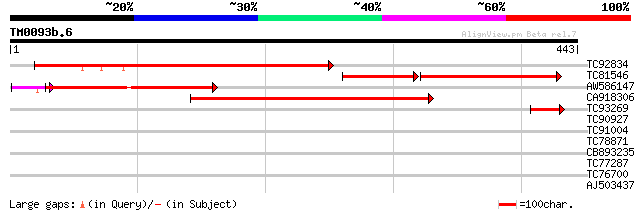
Score E
Sequences producing significant alignments: (bits) Value
TC92834 similar to GP|16209669|gb|AAL14393.1 AT4g02500/T10P11_20... 276 1e-74
TC81546 similar to GP|16209669|gb|AAL14393.1 AT4g02500/T10P11_20... 174 5e-68
AW586147 similar to PIR|E96772|E96 hypothetical protein F1M20.6 ... 241 3e-66
CA918306 similar to PIR|T04726|T04 hypothetical protein F19F18.1... 227 6e-60
TC93269 similar to PIR|E96772|E96772 hypothetical protein F1M20.... 55 7e-08
TC90927 similar to GP|9279712|dbj|BAB01269.1 cell division prote... 29 3.2
TC91004 28 5.4
TC78871 similar to GP|23180022|gb|AAN14398.1 CG31065-PA {Drosoph... 28 7.1
CB893235 similar to GP|6625560|gb|A beta-glucan binding protein ... 28 7.1
TC77287 SP|Q02735|CAPP_MEDSA Phosphoenolpyruvate carboxylase (EC... 28 9.3
TC76700 SP|O48905|MDHC_MEDSA Malate dehydrogenase cytoplasmic (... 28 9.3
AJ503437 similar to PIR|E96772|E967 hypothetical protein F1M20.6... 28 9.3
>TC92834 similar to GP|16209669|gb|AAL14393.1 AT4g02500/T10P11_20
{Arabidopsis thaliana}, partial (46%)
Length = 910
Score = 276 bits (705), Expect = 1e-74
Identities = 130/256 (50%), Positives = 177/256 (68%), Gaps = 22/256 (8%)
Frame = +1
Query: 20 GGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVN---LSSSDADAVNQNVIE-----E 71
G RR RQ+Q+ F +T LC F+T++VLRGTIG D + + Q + E
Sbjct: 142 GSRRLRQMQRAFRRGTLTFLCLFLTVIVLRGTIGAGKFGTPEQDLNEIRQQLYSRGRRVE 321
Query: 72 TNRILAEIRSDADPSD-------------PDDAAAAETFFSPNATFTLGPKITGWDLQRK 118
+R+L E++S + +D D+ A + PN +TLGPKI+ WD QR
Sbjct: 322 PHRVLEEVQSSENNNDNNNYATFDITKILKDEEAGDDEKRDPNTPYTLGPKISDWDEQRS 501
Query: 119 AWLDQNPEYPNFVR-GKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVY 177
+WL NP+YPNF+ K R+LL+TGS PKP +NP+GDHYLLKSIKNKIDYCRLHGIEI Y
Sbjct: 502 SWLSNNPDYPNFINPNKPRVLLITGSSPKPGENPVGDHYLLKSIKNKIDYCRLHGIEIFY 681
Query: 178 NLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVL 237
N+A LD E+AG+WAKLP+IR+L+LSHPEVE++WWMDSDA FTDM FE+P +Y D+N V+
Sbjct: 682 NMALLDAEMAGFWAKLPLIRKLLLSHPEVEFLWWMDSDAMFTDMAFEVPWERYKDHNFVM 861
Query: 238 HGYPDLLFEQKSWIAV 253
HG+ ++++++K+WI +
Sbjct: 862 HGWNEMIYDEKNWIGL 909
>TC81546 similar to GP|16209669|gb|AAL14393.1 AT4g02500/T10P11_20
{Arabidopsis thaliana}, partial (39%)
Length = 816
Score = 174 bits (442), Expect(2) = 5e-68
Identities = 71/110 (64%), Positives = 88/110 (79%)
Frame = +3
Query: 322 KTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDERWPFVTHFVGCKPCGSYGDYPVEKC 381
K +LEN +YLHGYW LVDRYEEMIE YHPG GD RWP VTHFVGCKPCG +GDYPVE+C
Sbjct: 189 KVYLENHYYLHGYWGILVDRYEEMIENYHPGFGDHRWPLVTHFVGCKPCGKFGDYPVERC 368
Query: 382 LSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNETVTPLEFVDQFDI 431
L M+RA+NF DNQ+L++YGF H+ L S ++KR+RNE+ PL+ D+ +
Sbjct: 369 LKQMDRAYNFGDNQILQMYGFTHKSLASRRVKRVRNESSNPLDVKDELGL 518
Score = 101 bits (252), Expect(2) = 5e-68
Identities = 42/59 (71%), Positives = 52/59 (87%)
Frame = +2
Query: 261 RNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSALIYLLLSKKDKW 319
RNCQWSLD+LDAWAPMGPKG +R+EAGK+LT LK RP FEADDQSA++YLL + +++W
Sbjct: 5 RNCQWSLDILDAWAPMGPKGKIRDEAGKILTRELKNRPVFEADDQSAMVYLLATGREQW 181
>AW586147 similar to PIR|E96772|E96 hypothetical protein F1M20.6 [imported] -
Arabidopsis thaliana, partial (28%)
Length = 534
Score = 241 bits (616), Expect(2) = 3e-66
Identities = 118/134 (88%), Positives = 125/134 (93%)
Frame = +3
Query: 29 KTFNNVKITILCGFVTILVLRGTIGVNLSSSDADAVNQNVIEETNRILAEIRSDADPSDP 88
KTFNNVKITILCGFVTILVLRGTIGVNL SSD+DAVNQN+IEETNRI+AEIRSDADPSDP
Sbjct: 138 KTFNNVKITILCGFVTILVLRGTIGVNLISSDSDAVNQNMIEETNRIIAEIRSDADPSDP 317
Query: 89 DDAAAAETFFSPNATFTLGPKITGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPC 148
DD TFF+PNATFTLGPKI+ WD RKAWL+QNPEYP+FVRGKARILLLTGSPPKPC
Sbjct: 318 DDKD--NTFFNPNATFTLGPKISDWDSDRKAWLNQNPEYPSFVRGKARILLLTGSPPKPC 491
Query: 149 DNPIGDHYLLKSIK 162
DNPIGDHYLLKSIK
Sbjct: 492 DNPIGDHYLLKSIK 533
Score = 28.9 bits (63), Expect(2) = 3e-66
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 4/38 (10%)
Frame = +1
Query: 2 NGSAHKRTTTGLPTTTARG----GRRGRQIQKTFNNVK 35
NGS H R+++ + TTA RRGRQI K +K
Sbjct: 46 NGSLHNRSSSAVLPTTATNSNSKNRRGRQISKHSTTLK 159
>CA918306 similar to PIR|T04726|T04 hypothetical protein F19F18.180 -
Arabidopsis thaliana, partial (39%)
Length = 635
Score = 227 bits (579), Expect = 6e-60
Identities = 99/190 (52%), Positives = 134/190 (70%)
Frame = +1
Query: 142 GSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLAHLDVELAGYWAKLPMIRRLML 201
GS P PC NPIGDH LL+ KNK+DY R+HG +I YN A L ++ YWAK P+++ M+
Sbjct: 4 GSQPSPCKNPIGDHLLLRFFKNKVDYSRIHGYDIFYNNALLHPKMFAYWAKYPVVKAAMM 183
Query: 202 SHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQKSWIAVNTGSFLFR 261
+HPE EWIWW+DSDA FTDM F+LPL +Y D+NLV+HG+P L+ E++SW +N G FL R
Sbjct: 184 AHPEAEWIWWVDSDALFTDMDFKLPLKRYKDHNLVVHGWPHLIHEKRSWTGLNAGVFLIR 363
Query: 262 NCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSALIYLLLSKKDKWMD 321
NCQWSLD ++AWA MGP+ P EE GK L + K + E+DDQ+ L YL+ +K+ +
Sbjct: 364 NCQWSLDFMEAWAGMGPQSPDYEEWGKTLRSTFKDKFFPESDDQTGLAYLIAIEKENGVT 543
Query: 322 KTFLENSFYL 331
+ + +YL
Sbjct: 544 RII*KGKYYL 573
Score = 30.0 bits (66), Expect = 1.9
Identities = 14/28 (50%), Positives = 16/28 (57%), Gaps = 3/28 (10%)
Frame = +3
Query: 311 LLLSKKD---KWMDKTFLENSFYLHGYW 335
L LS +D KW DK +LE L GYW
Sbjct: 501 LSLSDRDRERKWGDKNYLEGEILLEGYW 584
>TC93269 similar to PIR|E96772|E96772 hypothetical protein F1M20.6
[imported] - Arabidopsis thaliana, partial (5%)
Length = 547
Score = 54.7 bits (130), Expect = 7e-08
Identities = 25/26 (96%), Positives = 26/26 (99%)
Frame = +3
Query: 408 LSPKIKRIRNETVTPLEFVDQFDIRR 433
LSPKIKRIRNET+TPLEFVDQFDIRR
Sbjct: 9 LSPKIKRIRNETITPLEFVDQFDIRR 86
>TC90927 similar to GP|9279712|dbj|BAB01269.1 cell division protein
FtsH-like {Arabidopsis thaliana}, partial (28%)
Length = 984
Score = 29.3 bits (64), Expect = 3.2
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 1/77 (1%)
Frame = -3
Query: 363 HFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNETVTP 422
HF+ C C ++K L S ++ ++K+YG +HR P I + + +
Sbjct: 475 HFISCTGC-ELNSNMIKKGLPS--------EHSIVKIYGLQHR----PNISQFPTSSRSN 335
Query: 423 LEFVDQFDIRR-HSSES 438
+ D +IR+ HSS S
Sbjct: 334 CDIFDIVEIRQVHSSNS 284
>TC91004
Length = 751
Score = 28.5 bits (62), Expect = 5.4
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = -2
Query: 322 KTFLENSFYLHGYWAGLVDRYEEMIEKYH 350
K L SF+L YW ++R EE +K H
Sbjct: 384 KQLLRQSFFLSSYWIKRIEREEERGKKTH 298
>TC78871 similar to GP|23180022|gb|AAN14398.1 CG31065-PA {Drosophila
melanogaster}, partial (3%)
Length = 1475
Score = 28.1 bits (61), Expect = 7.1
Identities = 11/24 (45%), Positives = 15/24 (61%)
Frame = +1
Query: 209 IWWMDSDAFFTDMVFELPLSKYDD 232
IW+M +FF D F L + +YDD
Sbjct: 229 IWFMIYFSFFIDFSFNLKIKRYDD 300
>CB893235 similar to GP|6625560|gb|A beta-glucan binding protein {Phaseolus
vulgaris}, partial (14%)
Length = 283
Score = 28.1 bits (61), Expect = 7.1
Identities = 10/25 (40%), Positives = 14/25 (56%)
Frame = +2
Query: 317 DKWMDKTFLENSFYLHGYWAGLVDR 341
+ W+D TF N F G W G+V +
Sbjct: 143 EPWLDGTFNGNGFLYDGKWGGIVTK 217
>TC77287 SP|Q02735|CAPP_MEDSA Phosphoenolpyruvate carboxylase (EC 4.1.1.31)
(PEPCASE). [Alfalfa] {Medicago sativa}, partial (33%)
Length = 1124
Score = 27.7 bits (60), Expect = 9.3
Identities = 14/54 (25%), Positives = 21/54 (37%), Gaps = 12/54 (22%)
Frame = -3
Query: 208 WIWWMDSDAFFTDMVFELPLSKYDDYNLVLHG------------YPDLLFEQKS 249
W WW D T+ LPL + L++ YP +L +Q+S
Sbjct: 813 WSWWSPLDFISTECSLNLPLESFIKLLLIIRSDVFSVQLR*TVPYPSVLLKQRS 652
>TC76700 SP|O48905|MDHC_MEDSA Malate dehydrogenase cytoplasmic (EC
1.1.1.37). [Alfalfa] {Medicago sativa}, complete
Length = 1413
Score = 27.7 bits (60), Expect = 9.3
Identities = 18/57 (31%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Frame = +3
Query: 195 MIRRLMLSH-PEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQKSW 250
+I L SH E++W WWM F ++ +L LS++ +++L D E+K W
Sbjct: 225 LILLLQQSH*MELKWSWWMLHFHFLKVLLLQLMLSRHALESILLSWLVDSQ-EKKVW 392
>AJ503437 similar to PIR|E96772|E967 hypothetical protein F1M20.6 [imported]
- Arabidopsis thaliana, partial (3%)
Length = 435
Score = 27.7 bits (60), Expect = 9.3
Identities = 16/29 (55%), Positives = 22/29 (75%), Gaps = 1/29 (3%)
Frame = +1
Query: 32 NNVK-ITILCGFVTILVLRGTIGVNLSSS 59
NN++ TIL GFVTILVL +G+N S++
Sbjct: 241 NNIQHSTILWGFVTILVLPCIMGLNDSTT 327
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,213,349
Number of Sequences: 36976
Number of extensions: 236979
Number of successful extensions: 1029
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1016
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1025
length of query: 443
length of database: 9,014,727
effective HSP length: 99
effective length of query: 344
effective length of database: 5,354,103
effective search space: 1841811432
effective search space used: 1841811432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0093b.6


