

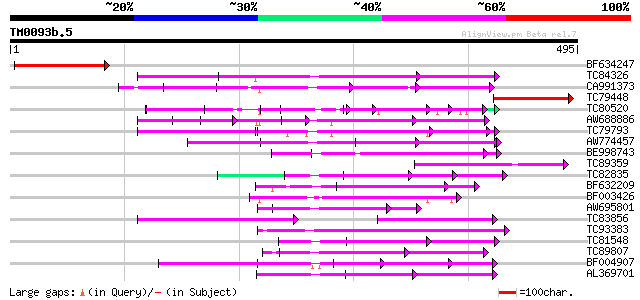
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093b.5
(495 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF634247 weakly similar to GP|21593376|gb| unknown {Arabidopsis ... 112 3e-25
TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein... 89 5e-18
CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imp... 84 1e-16
TC79448 similar to GP|12321513|gb|AAG50816.1 unknown protein {Ar... 82 6e-16
TC80520 weakly similar to PIR|T02562|T02562 probable salt-induci... 60 7e-16
AW688886 similar to GP|3258568|gb|A Unknown protein {Arabidopsis... 81 8e-16
TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Ar... 80 1e-15
AW774457 weakly similar to GP|10176973|dbj gb|AAF19552.1~gene_id... 76 3e-14
BE998743 similar to GP|22128591|gb| fertility restorer-like prot... 68 9e-12
TC89359 weakly similar to GP|10177319|dbj|BAB10645. gb|AAF26800.... 68 9e-12
TC82835 weakly similar to GP|21741712|emb|CAD41335. oj991113_30.... 66 3e-11
BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01... 62 5e-10
BF003426 similar to PIR|F96760|F96 hypothetical protein T9L24.39... 62 5e-10
AW695801 weakly similar to PIR|T01622|T01 probable salt-inducibl... 62 5e-10
TC83856 weakly similar to PIR|B96656|B96656 unknown protein 419... 59 3e-09
TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16... 56 3e-08
TC81548 weakly similar to GP|1931651|gb|AAB65486.1| membrane-ass... 55 5e-08
TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [i... 55 5e-08
BF004907 weakly similar to GP|8953393|emb putative protein {Arab... 55 6e-08
AL369701 weakly similar to PIR|E84433|E8 hypothetical protein At... 54 1e-07
>BF634247 weakly similar to GP|21593376|gb| unknown {Arabidopsis thaliana},
partial (7%)
Length = 345
Score = 112 bits (280), Expect = 3e-25
Identities = 60/83 (72%), Positives = 65/83 (78%)
Frame = +1
Query: 5 MLNSAWKLCCLRKTRTRNFQLLSASLCSTLQPISAPPHLQELCGIVTSNVGGLDDLELSL 64
+LNSA+K L+ T T FQLLS SL STL PIS PP LQ+LC IVTS VGGLDDLE SL
Sbjct: 94 LLNSAFKRYWLQNTSTHKFQLLSVSLFSTLHPISTPPLLQDLCDIVTSTVGGLDDLESSL 273
Query: 65 NKFKGSLTSSLVAQAIESSKHEA 87
NKFKGSLTS LVAQ I+S KHEA
Sbjct: 274 NKFKGSLTSPLVAQVIDSVKHEA 342
>TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein AT4g19900
[imported] - Arabidopsis thaliana, partial (18%)
Length = 756
Score = 88.6 bits (218), Expect = 5e-18
Identities = 58/245 (23%), Positives = 107/245 (43%)
Frame = +1
Query: 183 TAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKS 242
TA+I C + RAE ++ K++ Y +L+ G N + A ++ M S
Sbjct: 4 TAMIRGYCREDKLNRAEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMSS 183
Query: 243 NGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLG 302
G P+L YN + LC+R G V E ++ + + + P +YNIL+S
Sbjct: 184 EGFSPNLCTYNAIVNGLCKR-------GRVQEAYKMLEDGFQNGLKPDKFTYNILMSEHC 342
Query: 303 KTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHK 362
K ++++ + M G+ PD SY ++ V R + + ++ + G++P +K
Sbjct: 343 KQENIRQALALFNKMLKIGIQPDIHSYTTLIAVFCRENRMKESEMFFEEAVRIGIIPTNK 522
Query: 363 FYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEA 422
Y S+I C + A++ F ++ Y +I LC+ ++ R L+D
Sbjct: 523 TYTSMICGYCREGNLTLAMKFFHRLSDHGCAPESITYGAIISGLCKQSKRDEARSLYDSM 702
Query: 423 TSMGI 427
G+
Sbjct: 703 IEKGL 717
Score = 75.1 bits (183), Expect = 6e-14
Identities = 50/251 (19%), Positives = 104/251 (40%), Gaps = 3/251 (1%)
Frame = +1
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
Y +R + + ++L+ +K++G V + T+ + + K G + A + +
Sbjct: 1 YTAMIRGYCREDKLNRAEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMS 180
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGAL---PCIYRSLLYGWSVHR 228
S N T AI+N LC +G + A ++ +D L Y L+
Sbjct: 181 SEGFSPNLCTYNAIVNGLCKRGRVQEAYKML---EDGFQNGLKPDKFTYNILMSEHCKQE 351
Query: 229 NVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVL 288
N+++A + +M G+ PD+ Y T + C N E + + ++
Sbjct: 352 NIRQALALFNKMLKIGIQPDIHSYTTLIAVFCRENRMKESEMFFEEAVRI-------GII 510
Query: 289 PTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEI 348
PT+ +Y ++ + + + + + G +P+ ++Y ++ L + + + +
Sbjct: 511 PTNKTYTSMICGYCREGNLTLAMKFFHRLSDHGCAPESITYGAIISGLCKQSKRDEARSL 690
Query: 349 VDQMIGKGLVP 359
D MI KGLVP
Sbjct: 691 YDSMIEKGLVP 723
Score = 33.1 bits (74), Expect = 0.25
Identities = 33/153 (21%), Positives = 63/153 (40%), Gaps = 4/153 (2%)
Frame = +1
Query: 67 FKGSLTSSLVAQAIESSKH--EAQTRRLLRFFLWSSKNLRHDLEDNDYNYA--LRVFAEK 122
F+ L I S+H + R+ L F +K L+ ++ + ++Y + VF +
Sbjct: 283 FQNGLKPDKFTYNILMSEHCKQENIRQALALF---NKMLKIGIQPDIHSYTTLIAVFCRE 453
Query: 123 QDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTV 182
++ + + G + +T+ + + G A+ F L + C+ T
Sbjct: 454 NRMKESEMFFEEAVRIGIIPTNKTYTSMICGYCREGNLTLAMKFFHRLSDHGCAPESITY 633
Query: 183 TAIINALCSKGHAKRAEGVVLHHKDKITGALPC 215
AII+ LC + +KR E L+ G +PC
Sbjct: 634 GAIISGLCKQ--SKRDEARSLYDSMIEKGLVPC 726
>CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 756
Score = 84.3 bits (207), Expect = 1e-16
Identities = 62/245 (25%), Positives = 110/245 (44%), Gaps = 2/245 (0%)
Frame = +2
Query: 181 TVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCI--YRSLLYGWSVHRNVKEARRIIK 238
T ++++ C +A+ ++ + G P I Y L+ G+ + V EA + K
Sbjct: 32 TYCSLMDGYCLVKEVNKAKSILYTMSQR--GVNPDIQSYNILIDGFCKIKKVDEAMNLFK 205
Query: 239 EMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILL 298
EM +IPD+V YN+ + LC+ G + L ++ EM V P I+Y+ +L
Sbjct: 206 EMHHKHIIPDVVTYNSLIDGLCK-------LGKISYALKLVDEMHDRGVPPDIITYSSIL 364
Query: 299 SCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLV 358
L K +V ++ +L +K G+ P+ +Y +++ L GR I + ++ KG
Sbjct: 365 DALCKNHQVDKAIALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYN 544
Query: 359 PNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGREL 418
Y +I C + AL L KMK +S Y+++I L + +K +L
Sbjct: 545 ITVNTYTVMIHGFCNKGLFDEALTLLSKMKDNSCIPDAVTYEIIIRSLFDKDENDKAEKL 724
Query: 419 WDEAT 423
+ T
Sbjct: 725 REMIT 739
Score = 68.2 bits (165), Expect = 7e-12
Identities = 52/226 (23%), Positives = 103/226 (45%), Gaps = 2/226 (0%)
Frame = +2
Query: 135 LKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGH 194
+ + G D Q++ ++ + K+ K DEA+ +FK + + T ++I+ LC G
Sbjct: 104 MSQRGVNPDIQSYNILIDGFCKIKKVDEAMNLFKEMHHKHIIPDVVTYNSLIDGLCKLGK 283
Query: 195 AKRAEGVVLHHKDKITGALPCI--YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCY 252
A +V D+ G P I Y S+L + V +A ++ ++K G+ P++ Y
Sbjct: 284 ISYALKLVDEMHDR--GVPPDIITYSSILDALCKNHQVDKAIALLTKLKDQGIRPNMYTY 457
Query: 253 NTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQ 312
+ LC+ G + + N+ ++ T +Y +++ E+
Sbjct: 458 TILIDGLCK-------GGRLEDAHNIFEDLLVKGYNITVNTYTVMIHGFCNKGLFDEALT 616
Query: 313 ILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLV 358
+L MK + PD V+Y +++R LF K +++ +MI +GL+
Sbjct: 617 LLSKMKDNSCIPDAVTYEIIIRSLFDKDENDKAEKL-REMITRGLL 751
Score = 52.8 bits (125), Expect = 3e-07
Identities = 40/206 (19%), Positives = 87/206 (41%)
Frame = +2
Query: 96 FLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLI 155
+ S + + D++ YN + F + + L ++ + + D T+ + + L
Sbjct: 98 YTMSQRGVNPDIQS--YNILIDGFCKIKKVDEAMNLFKEMHHKHIIPDVVTYNSLIDGLC 271
Query: 156 KLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPC 215
KLGK AL + + + T ++I++ALC +A ++ KD+
Sbjct: 272 KLGKISYALKLVDEMHDRGVPPDIITYSSILDALCKNHQVDKAIALLTKLKDQGIRPNMY 451
Query: 216 IYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPET 275
Y L+ G +++A I +++ G + Y + C + GL E
Sbjct: 452 TYTILIDGLCKGGRLEDAHNIFEDLLVKGYNITVNTYTVMIHGFCNK-------GLFDEA 610
Query: 276 LNVMMEMRSHKVLPTSISYNILLSCL 301
L ++ +M+ + +P +++Y I++ L
Sbjct: 611 LTLLSKMKDNSCIPDAVTYEIIIRSL 688
>TC79448 similar to GP|12321513|gb|AAG50816.1 unknown protein {Arabidopsis
thaliana}, partial (10%)
Length = 1019
Score = 81.6 bits (200), Expect = 6e-16
Identities = 41/70 (58%), Positives = 50/70 (70%)
Frame = +1
Query: 423 TSMGITLQCSKDILDPSITEVFKPVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAV 482
T MGITLQCSKD+LDPSITEV+ P RPEKI++ DSP AKS +K ++KMRK
Sbjct: 4 TLMGITLQCSKDVLDPSITEVYIPKRPEKINVVDSPKAKSQQKLSNYFERMKMRKAAAWK 183
Query: 483 KKKKKRQKKS 492
KK KK+ + S
Sbjct: 184 KKTKKKSEAS 213
>TC80520 weakly similar to PIR|T02562|T02562 probable salt-inducible protein
[imported] - Arabidopsis thaliana, partial (6%)
Length = 1911
Score = 59.7 bits (143), Expect = 3e-09
Identities = 49/218 (22%), Positives = 97/218 (44%), Gaps = 1/218 (0%)
Frame = +3
Query: 171 DKYKCSTNEFT-VTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRN 229
D + C +F + + +C +G+A E VV Y +++ G +H
Sbjct: 1020 DIWYCKVRDFAEARKLYDDMCGRGYA---ENVVS-------------YSTMISGLYLHGK 1151
Query: 230 VKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLP 289
EA + EM G+ DL+ YN+ ++ LC+ G + + N++ ++ + P
Sbjct: 1152 TDEALSLFHEMSRKGIARDLISYNSLIKGLCQ-------EGELAKATNLLNKLLIQGLEP 1310
Query: 290 TSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIV 349
+ S+ L+ CL K + + ++L+ M + P ++ ++ L G F +G E +
Sbjct: 1311 SVSSFTPLIKCLCKVGDTEGAMRLLKDMHDRHLEPIASTHDYMIIGLSKKGDFAQGMEWL 1490
Query: 350 DQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKM 387
+M+ L PN + + LI L R++ L + + M
Sbjct: 1491 LKMLSWKLKPNMQTFEHLIDCLKRENRLDDILIVLDLM 1604
Score = 58.5 bits (140), Expect(2) = 7e-16
Identities = 33/126 (26%), Positives = 61/126 (48%)
Frame = +3
Query: 292 ISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQ 351
+SY+ ++S L + E+ + M G++ D +SY +++ L G K ++++
Sbjct: 1107 VSYSTMISGLYLHGKTDEALSLFHEMSRKGIARDLISYNSLIKGLCQEGELAKATNLLNK 1286
Query: 352 MIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGD 411
++ +GL P+ + LI LC V A+ L + M L +D +I L + GD
Sbjct: 1287 LLIQGLEPSVSSFTPLIKCLCKVGDTEGAMRLLKDMHDRHLEPIASTHDYMIIGLSKKGD 1466
Query: 412 FEKGRE 417
F +G E
Sbjct: 1467 FAQGME 1484
Score = 53.1 bits (126), Expect = 2e-07
Identities = 35/149 (23%), Positives = 70/149 (46%)
Frame = +1
Query: 220 LLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVM 279
L+ + V +++++ G+ D +N + C++ V E L++M
Sbjct: 598 LIKAFCAENKVFNGYELLRQVLEKGLCVDNTVFNALINGFCKQKQYDR----VSEILHIM 765
Query: 280 MEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLS 339
+ M+ + P+ +Y +++ L K R+ E+ ++ +K G PD V Y V++
Sbjct: 766 IAMKCN---PSIYTYQEIINGLLKRRKNDEAFRVFNDLKDRGYFPDRVMYTTVIKGFCDM 936
Query: 340 GRFGKGKEIVDQMIGKGLVPNHKFYFSLI 368
G + +++ +MI KGLVPN Y +I
Sbjct: 937 GLLAEARKLWFEMIQKGLVPNEYTYNVMI 1023
Score = 48.1 bits (113), Expect = 8e-06
Identities = 40/205 (19%), Positives = 90/205 (43%), Gaps = 2/205 (0%)
Frame = +3
Query: 119 FAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTN 178
+ + +D+ L D+ G + ++ + L GK DEAL +F + + + +
Sbjct: 1029 YCKVRDFAEARKLYDDMCGRGYAENVVSYSTMISGLYLHGKTDEALSLFHEMSRKGIARD 1208
Query: 179 EFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCI--YRSLLYGWSVHRNVKEARRI 236
+ ++I LC +G +A ++ +K I G P + + L+ + + A R+
Sbjct: 1209 LISYNSLIKGLCQEGELAKATNLL--NKLLIQGLEPSVSSFTPLIKCLCKVGDTEGAMRL 1382
Query: 237 IKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNI 296
+K+M + P ++ + L ++ G + + +++M S K+ P ++
Sbjct: 1383 LKDMHDRHLEPIASTHDYMIIGLSKK-------GDFAQGMEWLLKMLSWKLKPNMQTFEH 1541
Query: 297 LLSCLGKTRRVKESCQILEAMKTSG 321
L+ CL + R+ + +L+ M G
Sbjct: 1542 LIDCLKRENRLDDILIVLDLMFREG 1616
Score = 43.9 bits (102), Expect = 1e-04
Identities = 57/229 (24%), Positives = 93/229 (39%), Gaps = 38/229 (16%)
Frame = +1
Query: 237 IKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNI 296
+K KS PD V N L R L N G+V E +V + ++ LP++ S+N+
Sbjct: 316 VKAAKSLLEYPDFVPKNDSLEGYV-RLLGEN--GMVEEVFDVFVSLKKVGFLPSASSFNV 486
Query: 297 -LLSCLGKTRRVKESCQILEAMKTS--GVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMI 353
LL+CL K R ++ E M S GV+ D + +++ + G E++ Q++
Sbjct: 487 CLLACL-KVGRTDLVWKLYELMIESGVGVNIDVETVGCLIKAFCAENKVFNGYELLRQVL 663
Query: 354 GKGLVPNHKFYFSLIGILC---GVERVNHALELFEKMKSSSL------------------ 392
KGL ++ + +LI C +RV+ L + MK +
Sbjct: 664 EKGLCVDNTVFNALINGFCKQKQYDRVSEILHIMIAMKCNPSIYTYQEIINGLLKRRKND 843
Query: 393 -----------GGYGP---VYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
GY P +Y +I C G + R+LW E G+
Sbjct: 844 EAFRVFNDLKDRGYFPDRVMYTTVIKGFCDMGLLAEARKLWFEMIQKGL 990
Score = 43.1 bits (100), Expect(2) = 7e-16
Identities = 38/179 (21%), Positives = 65/179 (36%)
Frame = +1
Query: 120 AEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNE 179
AE + + ++L L+K G +D F + K + D I + KC+ +
Sbjct: 616 AENKVFNGYELLRQVLEK-GLCVDNTVFNALINGFCKQKQYDRVSEILHIMIAMKCNPSI 792
Query: 180 FTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIKE 239
+T IIN L + R EA R+ +
Sbjct: 793 YTYQEIINGLLKR-----------------------------------RKNDEAFRVFND 867
Query: 240 MKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILL 298
+K G PD V Y T ++ C+ GL+ E + EM ++P +YN+++
Sbjct: 868 LKDRGYFPDRVMYTTVIKGFCD-------MGLLAEARKLWFEMIQKGLVPNEYTYNVMI 1023
Score = 41.2 bits (95), Expect = 0.001
Identities = 32/116 (27%), Positives = 51/116 (43%)
Frame = +3
Query: 303 KTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHK 362
K R E+ ++ + M G + + VSY ++ L+L G+ + + +M KG+ +
Sbjct: 1035 KVRDFAEARKLYDDMCGRGYAENVVSYSTMISGLYLHGKTDEALSLFHEMSRKGIARDLI 1214
Query: 363 FYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGREL 418
Y SLI LC + A L K+ L + LI LC+ GD E L
Sbjct: 1215 SYNSLIKGLCQEGELAKATNLLNKLLIQGLEPSVSSFTPLIKCLCKVGDTEGAMRL 1382
>AW688886 similar to GP|3258568|gb|A Unknown protein {Arabidopsis thaliana},
partial (11%)
Length = 660
Score = 81.3 bits (199), Expect = 8e-16
Identities = 56/185 (30%), Positives = 92/185 (49%), Gaps = 3/185 (1%)
Frame = +2
Query: 238 KEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVM--MEMRSHKVLPTSISYN 295
KEM S PD+V YNT L +C G + N++ M + + P ++Y
Sbjct: 14 KEMTSFDCDPDVVTYNTALLMVCVE------XGKIKVAHNLVNGMSKKCKDLSPDVVTYT 175
Query: 296 ILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGK 355
L+ + + V E+ ILE M G+ P+ V+Y +++ L + ++ K KEI++QM G
Sbjct: 176 TLIRGYCRKQEVDEALDILEEMNGRGLKPNIVTYNTLIKGLCEAQKWDKMKEILEQMKGD 355
Query: 356 -GLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEK 414
G +P+ + +LI C ++ A ++FE MK + Y VLI LC+ GD+ K
Sbjct: 356 GGSIPDACTFNTLINSHCCAGNLDEAFKVFENMKKLEVSADSASYSVLIRTLCQKGDYGK 535
Query: 415 GRELW 419
L+
Sbjct: 536 AEMLF 550
Score = 66.2 bits (160), Expect = 3e-11
Identities = 50/194 (25%), Positives = 92/194 (46%), Gaps = 4/194 (2%)
Frame = +2
Query: 167 FKNLDKYKCSTNEFTV-TAIINALCSKGHAKRAEGVVLHHKDKITGALPCI--YRSLLYG 223
FK + + C + T TA++ G K A +V K P + Y +L+ G
Sbjct: 11 FKEMTSFDCDPDVVTYNTALLMVCVEXGKIKVAHNLVNGMSKKCKDLSPDVVTYTTLIRG 190
Query: 224 WSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMR 283
+ + V EA I++EM G+ P++V YNT ++ LCE + + ++ +M+
Sbjct: 191 YCRKQEVDEALDILEEMNGRGLKPNIVTYNTLIKGLCE-------AQKWDKMKEILEQMK 349
Query: 284 SH-KVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRF 342
+P + ++N L++ + E+ ++ E MK VS D SY +++R L G +
Sbjct: 350 GDGGSIPDACTFNTLINSHCCAGNLDEAFKVFENMKKLEVSADSASYSVLIRTLCQKGDY 529
Query: 343 GKGKEIVDQMIGKG 356
GK + + +I +G
Sbjct: 530 GKAEMLF*XVI*EG 571
Score = 47.4 bits (111), Expect = 1e-05
Identities = 26/89 (29%), Positives = 45/89 (50%), Gaps = 1/89 (1%)
Frame = +2
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEG-RVMDAQTFGLVAENLIKLGKEDEALGIFKNL 170
YN ++ E Q + M ++ +K +G + DA TF + + G DEA +F+N+
Sbjct: 275 YNTLIKGLCEAQKWDKMKEILEQMKGDGGSIPDACTFNTLINSHCCAGNLDEAFKVFENM 454
Query: 171 DKYKCSTNEFTVTAIINALCSKGHAKRAE 199
K + S + + + +I LC KG +AE
Sbjct: 455 KKLEVSADSASYSVLIRTLCQKGDYGKAE 541
Score = 41.6 bits (96), Expect = 7e-04
Identities = 28/122 (22%), Positives = 53/122 (42%), Gaps = 2/122 (1%)
Frame = +2
Query: 143 DAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVV 202
D T+ + + + DEAL I + ++ N T +I LC + + ++
Sbjct: 158 DVVTYTTLIRGYCRKQEVDEALDILEEMNGRGLKPNIVTYNTLIKGLCEAQKWDKMKEIL 337
Query: 203 LHHKDKITGALP--CIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLC 260
K G++P C + +L+ N+ EA ++ + MK V D Y+ +R LC
Sbjct: 338 EQMKGD-GGSIPDACTFNTLINSHCCAGNLDEAFKVFENMKKLEVSADSASYSVLIRTLC 514
Query: 261 ER 262
++
Sbjct: 515 QK 520
>TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Arabidopsis
thaliana}, partial (7%)
Length = 1580
Score = 80.5 bits (197), Expect = 1e-15
Identities = 53/211 (25%), Positives = 98/211 (46%)
Frame = +1
Query: 217 YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETL 276
Y ++ G+ + EA + KEM +IPD+V Y++ + L + SG + L
Sbjct: 670 YSIMINGFCKIKKFDEAMNLFKEMHRKNIIPDVVTYSSLIDGLSK-------SGRISYAL 828
Query: 277 NVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVL 336
++ +M V P +YN +L L KT +V ++ +L K G PD +Y ++++ L
Sbjct: 829 QLVDQMHDRGVPPNICTYNSILDALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGL 1008
Query: 337 FLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYG 396
SG+ +++ + ++ KG N Y +I C N AL L KM+ +
Sbjct: 1009CQSGKLEDARKVFEGLLVKGHNLNVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDA 1188
Query: 397 PVYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
Y+++I L + + + +L E + G+
Sbjct: 1189KTYEIIILSLFKKDENDMAEKLLREMIARGL 1281
Score = 78.6 bits (192), Expect = 5e-15
Identities = 57/212 (26%), Positives = 98/212 (45%), Gaps = 6/212 (2%)
Frame = +1
Query: 215 CIYRSLLYGWSVHRNVKEARRIIKEMK----SNGVIPDLVCYNTFLR--CLCERNLRHNP 268
C+Y LL W +H E + +K + G+ P+ V YN+ + CL +
Sbjct: 448 CVY--LLIYWLMHFVRTER*KKVKLCLI***NKGIKPNFVTYNSLMDGYCLVKE------ 603
Query: 269 SGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVS 328
V + ++ M V P SY+I+++ K ++ E+ + + M + PD V+
Sbjct: 604 ---VNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDEAMNLFKEMHRKNIIPDVVT 774
Query: 329 YYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMK 388
Y ++ L SGR ++VDQM +G+ PN Y S++ LC +V+ A+ L K K
Sbjct: 775 YSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDALCKTHQVDKAIALLTKFK 954
Query: 389 SSSLGGYGPVYDVLIPKLCRGGDFEKGRELWD 420
Y +LI LC+ G E R++++
Sbjct: 955 DKGFQPDISTYSILIKGLCQSGKLEDARKVFE 1050
Score = 69.3 bits (168), Expect = 3e-12
Identities = 55/264 (20%), Positives = 114/264 (42%), Gaps = 4/264 (1%)
Frame = +1
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
YN + + ++ + + + G D Q++ ++ K+ K DEA+ +FK +
Sbjct: 565 YNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDEAMNLFKEMH 744
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVK 231
+ + T +++I+ L G A +V D+ C Y S+L V
Sbjct: 745 RKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDALCKTHQVD 924
Query: 232 EARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVM--MEMRSHKVLP 289
+A ++ + K G PD+ Y+ ++ LC+ SG + + V + ++ H +
Sbjct: 925 KAIALLTKFKDKGFQPDISTYSILIKGLCQ-------SGKLEDARKVFEGLLVKGHNL-- 1077
Query: 290 TSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIV 349
+Y I++ E+ +L M+ +G PD +Y +++ LF ++++
Sbjct: 1078NVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEIIILSLFKKDENDMAEKLL 1257
Query: 350 DQMIGKGLVPNHKF--YFSLIGIL 371
+MI +GL N + YF + +L
Sbjct: 1258REMIARGLP*N*NYVRYFCIYTLL 1329
>AW774457 weakly similar to GP|10176973|dbj
gb|AAF19552.1~gene_id:MTG13.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (13%)
Length = 650
Score = 75.9 bits (185), Expect = 3e-14
Identities = 50/203 (24%), Positives = 98/203 (47%)
Frame = -1
Query: 156 KLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPC 215
K+ DEAL +F ++ + ++ T ++I+ LC G A +V D A
Sbjct: 632 KIKMVDEALSLFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDNGQPANIF 453
Query: 216 IYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPET 275
Y L+ + +V +A ++K++K G+ PD+ +N + LC+ G +
Sbjct: 452 TYNCLIDALCKNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCK-------VGRLKNA 294
Query: 276 LNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRV 335
+V ++ S + +YNI+++ L K E+ +L M +G+ PD V+Y +++
Sbjct: 293 QDVFQDLLSKGYSVNAWTYNIMVNGLCKEGLFDEAEALLSKMDDNGIIPDAVTYETLIQA 114
Query: 336 LFLSGRFGKGKEIVDQMIGKGLV 358
LF K ++++ +MI +GL+
Sbjct: 113 LFHKDENEKAEKLLREMIARGLL 45
Score = 75.5 bits (184), Expect = 4e-14
Identities = 49/208 (23%), Positives = 93/208 (44%)
Frame = -1
Query: 220 LLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVM 279
++ G+ + V EA + +M+ G+ PD V YN+ + LC+ SG + ++
Sbjct: 650 MINGFCKIKMVDEALSLFNDMQFKGIAPDKVTYNSLIDGLCK-------SGRISYAWELV 492
Query: 280 MEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLS 339
EM + +YN L+ L K V ++ +++ +K G+ PD ++ +++ L
Sbjct: 491 DEMHDNGQPANIFTYNCLIDALCKNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCKV 312
Query: 340 GRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVY 399
GR +++ ++ KG N Y ++ LC + A L KM + + Y
Sbjct: 311 GRLKNAQDVFQDLLSKGYSVNAWTYNIMVNGLCKEGLFDEAEALLSKMDDNGIIPDAVTY 132
Query: 400 DVLIPKLCRGGDFEKGRELWDEATSMGI 427
+ LI L + EK +L E + G+
Sbjct: 131 ETLIQALFHKDENEKAEKLLREMIARGL 48
Score = 60.8 bits (146), Expect = 1e-09
Identities = 36/127 (28%), Positives = 64/127 (50%)
Frame = -1
Query: 303 KTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHK 362
K + V E+ + M+ G++PD V+Y ++ L SGR E+VD+M G N
Sbjct: 632 KIKMVDEALSLFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDNGQPANIF 453
Query: 363 FYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEA 422
Y LI LC V+ A+ L +K+K + +++LI LC+ G + ++++ +
Sbjct: 452 TYNCLIDALCKNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCKVGRLKNAQDVFQDL 273
Query: 423 TSMGITL 429
S G ++
Sbjct: 272 LSKGYSV 252
>BE998743 similar to GP|22128591|gb| fertility restorer-like protein {Petunia
x hybrida}, partial (7%)
Length = 829
Score = 67.8 bits (164), Expect = 9e-12
Identities = 49/190 (25%), Positives = 90/190 (46%)
Frame = +2
Query: 229 NVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVL 288
NVK A+ + M + PD+V YN+ + C N V + +V+ + V
Sbjct: 116 NVKGAKNALAMMIKGSIKPDVVTYNSLMDGYCLVNE-------VNKAKHVLSTIARMGVA 274
Query: 289 PTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEI 348
P + SYNI+++ R+ + ++++ M +G PD +Y ++ L + K +
Sbjct: 275 PDAQSYNIMVNGFC---RISHAWKLVDEMHVNGQPPDIFTYSSLIDALCKNNHLDKAIAL 445
Query: 349 VDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCR 408
V ++ +G+ PN Y LI LC R+ +A ++F+ + + Y++LI LC+
Sbjct: 446 VKKIKDQGIQPNMYTYNILIDGLCKGGRLKNAQDVFQDLLTKGYSLNIRTYNILINGLCK 625
Query: 409 GGDFEKGREL 418
G F+K L
Sbjct: 626 EGLFDKAEAL 655
Score = 67.0 bits (162), Expect = 2e-11
Identities = 44/166 (26%), Positives = 77/166 (45%)
Frame = +2
Query: 264 LRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVS 323
LR G V N + M + P ++YN L+ V ++ +L + GV+
Sbjct: 95 LRMRKEGNVKGAKNALAMMIKGSIKPDVVTYNSLMDGYCLVNEVNKAKHVLSTIARMGVA 274
Query: 324 PDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALEL 383
PD SY ++V R ++VD+M G P+ Y SLI LC ++ A+ L
Sbjct: 275 PDAQSYNIMVNGFC---RISHAWKLVDEMHVNGQPPDIFTYSSLIDALCKNNHLDKAIAL 445
Query: 384 FEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGITL 429
+K+K + Y++LI LC+GG + ++++ + + G +L
Sbjct: 446 VKKIKDQGIQPNMYTYNILIDGLCKGGRLKNAQDVFQDLLTKGYSL 583
>TC89359 weakly similar to GP|10177319|dbj|BAB10645.
gb|AAF26800.1~gene_id:MSL3.8~similar to unknown protein
{Arabidopsis thaliana}, partial (23%)
Length = 662
Score = 67.8 bits (164), Expect = 9e-12
Identities = 41/135 (30%), Positives = 66/135 (48%)
Frame = +3
Query: 354 GKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFE 413
G G + K YF + ILCG+ER+NHAL++F MK+ YD+L+ KL +
Sbjct: 9 GYGAFLDKKAYFDFLTILCGIERINHALKIFTMMKADGCEPGVKTYDLLMKKLGAHDRVD 188
Query: 414 KGRELWDEATSMGITLQCSKDILDPSITEVFKPVRPEKISLADSPIAKSPKKAKKLLGKV 473
K L++EA S G+ + + ++DP F + +K A+ P+K + ++
Sbjct: 189 KANALFNEARSRGLDVTLKEHVVDPR----FSKKKEKKAVKAEKKRETLPEKMARKRRRL 356
Query: 474 KMRKKPTAVKKKKKR 488
K + K KK R
Sbjct: 357 KQIRLSFVKKPKKSR 401
>TC82835 weakly similar to GP|21741712|emb|CAD41335. oj991113_30.17 {Oryza
sativa}, partial (5%)
Length = 915
Score = 65.9 bits (159), Expect = 3e-11
Identities = 45/151 (29%), Positives = 74/151 (48%)
Frame = +3
Query: 241 KSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSC 300
+ N DL+ YN L L + G V E L++ ++ +P +ISYN L++
Sbjct: 90 EQNNSCLDLITYNIVLDILGRK-------GRVDEMLDMFASLKETGFVPDTISYNTLING 248
Query: 301 LGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPN 360
L K R + + MK +G PD ++Y ++ + +G + + +M KG++P+
Sbjct: 249 LRKVGRSDMCFEYFKEMKENGNEPDLLTYTALIDISGRAGNIEESLKFFMEMKLKGILPS 428
Query: 361 HKFYFSLIGILCGVERVNHALELFEKMKSSS 391
+ Y SLI L E + A EL E+M SSS
Sbjct: 429 IQIYRSLIHNLNKTENIELATELLEEMNSSS 521
Score = 57.0 bits (136), Expect = 2e-08
Identities = 42/143 (29%), Positives = 64/143 (44%)
Frame = +3
Query: 292 ISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQ 351
I+YNI+L LG+ RV E + ++K +G PD +SY ++ L GR E +
Sbjct: 117 ITYNIVLDILGRKGRVDEMLDMFASLKETGFVPDTISYNTLINGLRKVGRSDMCFEYFKE 296
Query: 352 MIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGD 411
M G P+ Y +LI I + +L+ F +MK + +Y LI L + +
Sbjct: 297 MKENGNEPDLLTYTALIDISGRAGNIEESLKFFMEMKLKGILPSIQIYRSLIHNLNKTEN 476
Query: 412 FEKGRELWDEATSMGITLQCSKD 434
E EL +E S L D
Sbjct: 477 IELATELLEEMNSSSTCLAVPGD 545
Score = 45.1 bits (105), Expect = 6e-05
Identities = 37/172 (21%), Positives = 65/172 (37%), Gaps = 1/172 (0%)
Frame = +3
Query: 182 VTAIINALCSKGHAKRAEGVVLHHKDKITGALPCI-YRSLLYGWSVHRNVKEARRIIKEM 240
+ II A G A + H +++ L I Y +L V E + +
Sbjct: 15 INKIIFAFAKSGQKDSALAIFEHLREQNNSCLDLITYNIVLDILGRKGRVDEMLDMFASL 194
Query: 241 KSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSC 300
K G +PD + YNT + L G EM+ + P ++Y L+
Sbjct: 195 KETGFVPDTISYNTLINGL-------RKVGRSDMCFEYFKEMKENGNEPDLLTYTALIDI 353
Query: 301 LGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQM 352
G+ ++ES + MK G+ P Y ++ L + E++++M
Sbjct: 354 SGRAGNIEESLKFFMEMKLKGILPSIQIYRSLIHNLNKTENIELATELLEEM 509
>BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01051 protein
domain and multiple PPR PF|01535 repeats. EST
gb|AA728420 comes, partial (13%)
Length = 508
Score = 62.0 bits (149), Expect = 5e-10
Identities = 38/125 (30%), Positives = 65/125 (51%)
Frame = +1
Query: 286 KVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKG 345
+V P + SY+I+++ K + V ++ + M+ ++PD V+Y ++ L SGR
Sbjct: 100 RVAPNARSYSIVINGFCKIKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYA 279
Query: 346 KEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPK 405
E+VD+M G + Y SLI LC V+ A+ L +K+K + Y++LI
Sbjct: 280 WELVDEMRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNILIDG 459
Query: 406 LCRGG 410
LC+ G
Sbjct: 460 LCKQG 474
Score = 53.9 bits (128), Expect = 1e-07
Identities = 41/175 (23%), Positives = 80/175 (45%), Gaps = 5/175 (2%)
Frame = +1
Query: 215 CIYRSLLYGWSVHR-----NVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPS 269
C Y + GW + + II M+ V P+ Y+ + C+ +
Sbjct: 7 CCYLQFINGWILSC**S**GQTCVKYIISRMR---VAPNARSYSIVINGFCKIKM----- 162
Query: 270 GLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSY 329
V + L++ EMR ++ P +++YN L+ L K+ R+ + ++++ M+ SG D ++Y
Sbjct: 163 --VDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYAWELVDEMRDSGQPADIITY 336
Query: 330 YLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELF 384
++ L + K +V ++ +G+ + Y LI LC R+N A +F
Sbjct: 337 NSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNILIDGLCKQGRLNDAQVIF 501
>BF003426 similar to PIR|F96760|F96 hypothetical protein T9L24.39 [imported]
- Arabidopsis thaliana, partial (43%)
Length = 774
Score = 62.0 bits (149), Expect = 5e-10
Identities = 53/205 (25%), Positives = 93/205 (44%), Gaps = 20/205 (9%)
Frame = +2
Query: 210 TGALPCI--YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHN 267
+G LP + Y+ ++ G + + EA + ++EM G PD+V +N FL+ LC HN
Sbjct: 29 SGCLPDVTTYKDIIEGMCLCGKIDEAYKFLEEMGKKGYPPDIVTHNCFLKVLC-----HN 193
Query: 268 PSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWV 327
E L + M +P+ +YN+L+S K + + M+ G PD
Sbjct: 194 KKS--EEALKLYGRMIELSCIPSVQTYNMLISMFFKMDDPDGAFETWHEMEKRGCRPDTD 367
Query: 328 SYYLVVRVLFLSGRFGKGKEIVDQMIGKGL-VPNHKF-----YFSLIGILCGVERVNHAL 381
+Y +++ LF + +++++I KG+ +P KF S IG L + +++ +
Sbjct: 368 TYGVMIEGLFNCNKAEDACILLEEVINKGIKLPYRKFDSLLMQLSEIGNLQAIHKLSDHM 547
Query: 382 ELF------------EKMKSSSLGG 394
F +K KS SL G
Sbjct: 548 RKFYNNPMARRFALSQKRKSMSLRG 622
Score = 32.3 bits (72), Expect = 0.43
Identities = 32/148 (21%), Positives = 56/148 (37%), Gaps = 2/148 (1%)
Frame = +2
Query: 132 IGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCS 191
+G + G + D T+ + E + GK DEA + + K + T + LC
Sbjct: 11 MGHMISSGCLPDVTTYKDIIEGMCLCGKIDEAYKFLEEMGKKGYPPDIVTHNCFLKVLCH 190
Query: 192 KGHAKRAEGVVLHHKDKITGALPCI--YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDL 249
+ K E + L+ + +P + Y L+ + + A EM+ G PD
Sbjct: 191 --NKKSEEALKLYGRMIELSCIPSVQTYNMLISMFFKMDDPDGAFETWHEMEKRGCRPDT 364
Query: 250 VCYNTFLRCLCERNLRHNPSGLVPETLN 277
Y + L N + L+ E +N
Sbjct: 365 DTYGVMIEGLFNCNKAEDACILLEEVIN 448
>AW695801 weakly similar to PIR|T01622|T01 probable salt-inducible protein
At2g18940 [imported] - Arabidopsis thaliana, partial
(18%)
Length = 655
Score = 62.0 bits (149), Expect = 5e-10
Identities = 31/117 (26%), Positives = 66/117 (55%)
Frame = +3
Query: 217 YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETL 276
Y SL+ ++ + +A ++K+++++G+ PD+V YNT ++ C++ GLV E +
Sbjct: 177 YNSLIDLYARVGDCWKAEEMLKDIQNSGISPDVVSYNTVIKGFCKK-------GLVQEAI 335
Query: 277 NVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVV 333
++ EM ++ V P I++N +SC E+ +++ M G P+ ++Y +V+
Sbjct: 336 RILSEMTANGVQPCPITFNTFMSCYAGNGLFAEADEVIRYMIEHGCMPNELTYKIVI 506
Score = 43.1 bits (100), Expect = 2e-04
Identities = 31/130 (23%), Positives = 61/130 (46%)
Frame = +3
Query: 230 VKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLP 289
+K R ++++NG D+V N+ L +R+ E L+V+ + P
Sbjct: 6 LKGMERAFHQLQNNGYKLDMVVINSMLSMF----VRNQKLEKAHEMLDVI---HVSGLQP 164
Query: 290 TSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIV 349
++YN L+ + ++ ++L+ ++ SG+SPD VSY V++ G + I+
Sbjct: 165 NLVTYNSLIDLYARVGDCWKAEEMLKDIQNSGISPDVVSYNTVIKGFCKKGLVQEAIRIL 344
Query: 350 DQMIGKGLVP 359
+M G+ P
Sbjct: 345 SEMTANGVQP 374
>TC83856 weakly similar to PIR|B96656|B96656 unknown protein 41955-40111
[imported] - Arabidopsis thaliana, partial (12%)
Length = 662
Score = 59.3 bits (142), Expect = 3e-09
Identities = 31/105 (29%), Positives = 52/105 (49%)
Frame = +2
Query: 322 VSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHAL 381
+ PD ++ ++V V SG+ ++VD+M +G PN Y S++ LC RV+ A+
Sbjct: 68 IKPDVYTFNILVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRVDKAV 247
Query: 382 ELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMG 426
L K+K + Y +LI LC G E R ++++ G
Sbjct: 248 ALLTKLKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKG 382
Score = 47.8 bits (112), Expect = 1e-05
Identities = 32/141 (22%), Positives = 59/141 (41%)
Frame = +2
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
+N + VF + + L+ ++ G+ + T+ + + L K + D+A+ + L
Sbjct: 89 FNILVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRVDKAVALLTKLK 268
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVK 231
N T T +I+ LC+ G + A + K Y + YG+
Sbjct: 269 DQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDITVVTYIVMFYGFCKKGLFD 448
Query: 232 EARRIIKEMKSNGVIPDLVCY 252
EA ++ +M+ NG IPD Y
Sbjct: 449 EASALLSKMEENGCIPDAKTY 511
>TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16.90 -
Arabidopsis thaliana, partial (43%)
Length = 1038
Score = 56.2 bits (134), Expect = 3e-08
Identities = 51/221 (23%), Positives = 90/221 (40%), Gaps = 1/221 (0%)
Frame = +1
Query: 217 YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETL 276
Y SL+ S N KE I +M+ + PD+V Y + + + E L
Sbjct: 13 YNSLM---SFETNYKEVSNIYDQMQRADLRPDVVSYALLINA-------YGKARREEEAL 162
Query: 277 NVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVL 336
V EM V PT +YNILL + V+++ + ++M+ PD SY ++
Sbjct: 163 AVFEEMLDAGVRPTRKAYNILLDAFSISGMVEQARIVFKSMRRDKYMPDLCSYTTMLSAY 342
Query: 337 FLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYG 396
+ ++ ++I G PN Y +LI + +E +E+M +
Sbjct: 343 VNAPDMEGAEKFFKRLIQDGFEPNVVTYGTLIKGYAKANDIEKVMEKYEEMLGRGIKANQ 522
Query: 397 PVYDVLIPKLCRGGDFEKGRELWDEATSMGITL-QCSKDIL 436
+ ++ + GDF+ + E G+ Q +K+IL
Sbjct: 523 TILTTIMDAHGKNGDFDSAVNWFKEMALNGLLPDQKAKNIL 645
Score = 38.9 bits (89), Expect = 0.005
Identities = 41/220 (18%), Positives = 85/220 (38%), Gaps = 29/220 (13%)
Frame = +1
Query: 124 DYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVT 183
+Y + + +++ D ++ L+ K +E+EAL +F+ +
Sbjct: 40 NYKEVSNIYDQMQRADLRPDVVSYALLINAYGKARREEEALAVFEEMLDAGVRPTRKAYN 219
Query: 184 AIINALCSKGHAKRAEGVVLH-HKDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKS 242
+++A G ++A V +DK L C Y ++L + +++ A + K +
Sbjct: 220 ILLDAFSISGMVEQARIVFKSMRRDKYMPDL-CSYTTMLSAYVNAPDMEGAEKFFKRLIQ 396
Query: 243 NGVIPDLVCYNTFLRCLCERNL----------------------------RHNPSGLVPE 274
+G P++V Y T ++ + N H +G
Sbjct: 397 DGFEPNVVTYGTLIKGYAKANDIEKVMEKYEEMLGRGIKANQTILTTIMDAHGKNGDFDS 576
Query: 275 TLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQIL 314
+N EM + +LP + NILLS +KE+ +++
Sbjct: 577 AVNWFKEMALNGLLPDQKAKNILLSLAKTEEDIKEANELV 696
>TC81548 weakly similar to GP|1931651|gb|AAB65486.1| membrane-associated
salt-inducible protein isolog; 88078-84012 {Arabidopsis
thaliana}, partial (23%)
Length = 882
Score = 55.5 bits (132), Expect = 5e-08
Identities = 32/135 (23%), Positives = 66/135 (48%), Gaps = 1/135 (0%)
Frame = +1
Query: 235 RIIKEMKSNGVIPDLVCYNTFLR-CLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSIS 293
++ ++MK +G++PDLV Y+T + C+ ++ P+ L ++ E++ +K+ +
Sbjct: 499 KLFRQMKHDGLVPDLVTYSTLIAGCVKVKD-------GYPKALELIQELQDNKLRMDDVI 657
Query: 294 YNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMI 353
Y +L+ + +E+ MK+ G SP+ Y ++ G F K ++ M
Sbjct: 658 YGAILAVCASNGKWEEAEYYFNQMKSEGRSPNVYHYSSLLNAYSACGDFTKADALIQDME 837
Query: 354 GKGLVPNHKFYFSLI 368
+GL PN +L+
Sbjct: 838 SEGLAPNKGILTTLL 882
Score = 42.0 bits (97), Expect = 5e-04
Identities = 32/134 (23%), Positives = 57/134 (41%), Gaps = 1/134 (0%)
Frame = +1
Query: 295 NILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYY-LVVRVLFLSGRFGKGKEIVDQMI 353
N +LSCL K + + ++ MK G+ PD V+Y L+ + + + K E++ ++
Sbjct: 448 NSVLSCLIKKGKFDTTMKLFRQMKHDGLVPDLVTYSTLIAGCVKVKDGYPKALELIQELQ 627
Query: 354 GKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFE 413
L + Y +++ + + A F +MKS Y L+ GDF
Sbjct: 628 DNKLRMDDVIYGAILAVCASNGKWEEAEYYFNQMKSEGRSPNVYHYSSLLNAYSACGDFT 807
Query: 414 KGRELWDEATSMGI 427
K L + S G+
Sbjct: 808 KADALIQDMESEGL 849
>TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [imported] -
Arabidopsis thaliana, partial (53%)
Length = 1241
Score = 55.5 bits (132), Expect = 5e-08
Identities = 46/199 (23%), Positives = 92/199 (46%), Gaps = 1/199 (0%)
Frame = +1
Query: 221 LYGWS-VHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVM 279
LYG S +HR+ A+++ EM ++ N L H+ V E L
Sbjct: 289 LYGKSNMHRH---AQKLFDEMPQRNCERSVLSLNALLAAYL-----HSKQYDVVERLFKK 444
Query: 280 MEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLS 339
+ ++ V P +SYN + L + + +LE M+ GV D +++ ++ L+
Sbjct: 445 LPVQL-SVKPDLVSYNTYIKALLEKGSFDSAVSVLEEMEKDGVESDLITFNTLLDGLYSK 621
Query: 340 GRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVY 399
GRF G+++ +++ K +VPN + Y + + L +R A+E +E+M+ + +
Sbjct: 622 GRFEDGEKLWEKLGEKNVVPNIRTYNARLLGLAVAKRAGEAVEFYEEMEKKGVKPDLFSF 801
Query: 400 DVLIPKLCRGGDFEKGREL 418
+ LI G+ ++ + +
Sbjct: 802 NALIKGFANEGNLDEAKNV 858
Score = 47.4 bits (111), Expect = 1e-05
Identities = 29/114 (25%), Positives = 53/114 (46%)
Frame = +1
Query: 236 IIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYN 295
+++EM+ +GV DL+ +NT L L + G + + ++ V+P +YN
Sbjct: 541 VLEEMEKDGVESDLITFNTLLDGLYSK-------GRFEDGEKLWEKLGEKNVVPNIRTYN 699
Query: 296 ILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIV 349
L L +R E+ + E M+ GV PD S+ +++ G + K +V
Sbjct: 700 ARLLGLAVAKRAGEAVEFYEEMEKKGVKPDLFSFNALIKGFANEGNLDEAKNVV 861
Score = 38.1 bits (87), Expect = 0.008
Identities = 30/129 (23%), Positives = 62/129 (47%), Gaps = 3/129 (2%)
Frame = +1
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
YN ++ EK + S ++ +++K+G D TF + + L G+ ++ +++ L
Sbjct: 484 YNTYIKALLEKGSFDSAVSVLEEMEKDGVESDLITFNTLLDGLYSKGRFEDGEKLWEKLG 663
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRA-EGVVLHHKDKITGALPCI--YRSLLYGWSVHR 228
+ N T A + L AKRA E V + + + G P + + +L+ G++
Sbjct: 664 EKNVVPNIRTYNARLLGLAV---AKRAGEAVEFYEEMEKKGVKPDLFSFNALIKGFANEG 834
Query: 229 NVKEARRII 237
N+ EA+ ++
Sbjct: 835 NLDEAKNVV 861
>BF004907 weakly similar to GP|8953393|emb putative protein {Arabidopsis
thaliana}, partial (34%)
Length = 665
Score = 55.1 bits (131), Expect = 6e-08
Identities = 44/199 (22%), Positives = 87/199 (43%), Gaps = 1/199 (0%)
Frame = +3
Query: 131 LIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALC 190
++G++ KEG +A + + + L + G+ EALG+ + + T +++ C
Sbjct: 93 MVGEMTKEGIKPNAIVYNPIIDALAEAGRFKEALGMMERFHVLQIGPTLSTYNSLVKGFC 272
Query: 191 SKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLV 250
G + A ++ + +P Y +S V E + +M +G PD +
Sbjct: 273 KAGDIEGASKILKKMISRGFLPIPTTYNYFFRYFSRCGKVDEGMNLYTKMIESGHNPDRL 452
Query: 251 CYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLS-CLGKTRRVKE 309
Y+ L+ LCE + + V MEMR HK ++ + +L+ L K +++E
Sbjct: 453 TYHLVLKMLCEEE-------KLELAVQVSMEMR-HKGYDMDLATSTMLTHLLCKMHKLEE 608
Query: 310 SCQILEAMKTSGVSPDWVS 328
+ E M G+ P +++
Sbjct: 609 AFAEFEDMIRRGIIPQYLT 665
Score = 53.9 bits (128), Expect = 1e-07
Identities = 44/199 (22%), Positives = 81/199 (40%), Gaps = 28/199 (14%)
Frame = +3
Query: 217 YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERN------------- 263
Y +L+ G+ R V++A ++ EM G+ P+ + YN + L E
Sbjct: 36 YGTLVEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRFKEALGMMERFH 215
Query: 264 -LRHNPS--------------GLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVK 308
L+ P+ G + ++ +M S LP +YN + +V
Sbjct: 216 VLQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYFFRYFSRCGKVD 395
Query: 309 ESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLI 368
E + M SG +PD ++Y+LV+++L + ++ +M KG + L
Sbjct: 396 EGMNLYTKMIESGHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMRHKGYDMDLATSTMLT 575
Query: 369 GILCGVERVNHALELFEKM 387
+LC + ++ A FE M
Sbjct: 576 HLLCKMHKLEEAFAEFEDM 632
Score = 48.1 bits (113), Expect = 8e-06
Identities = 30/144 (20%), Positives = 65/144 (44%)
Frame = +3
Query: 283 RSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRF 342
++ V P+ ++Y L+ + RRV+++ +++ M G+ P+ + Y ++ L +GRF
Sbjct: 3 KNENVRPSVVTYGTLVEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRF 182
Query: 343 GKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVL 402
+ ++++ + P Y SL+ C + A ++ +KM S Y+
Sbjct: 183 KEALGMMERFHVLQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYF 362
Query: 403 IPKLCRGGDFEKGRELWDEATSMG 426
R G ++G L+ + G
Sbjct: 363 FRYFSRCGKVDEGMNLYTKMIESG 434
>AL369701 weakly similar to PIR|E84433|E8 hypothetical protein At2g02150
[imported] - Arabidopsis thaliana, partial (11%)
Length = 463
Score = 53.9 bits (128), Expect = 1e-07
Identities = 37/141 (26%), Positives = 64/141 (45%)
Frame = +1
Query: 216 IYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPET 275
IY SL++G+ + +++A I+KEM D Y T + LC +N + E+
Sbjct: 61 IYTSLIHGYMKAKMMEKAMDILKEMNKKNFKLDSPLYGTKIWGLCSQN-------KIEES 219
Query: 276 LNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRV 335
VM EM+ H + S Y L+ K ++ E+ +L+ M+ G+ V+Y +++
Sbjct: 220 EAVMREMKDHGLTANSYIYTSLMDAYFKVGKITEAVNLLQEMQKFGIETTAVTYGVLIDG 399
Query: 336 LFLSGRFGKGKEIVDQMIGKG 356
L G + D M G
Sbjct: 400 LCKKGLVQQAVSYFDCMTKTG 462
Score = 42.4 bits (98), Expect = 4e-04
Identities = 30/133 (22%), Positives = 56/133 (41%)
Frame = +1
Query: 294 YNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMI 353
Y L+ K + ++++ IL+ M D Y + L + + + ++ +M
Sbjct: 64 YTSLIHGYMKAKMMEKAMDILKEMNKKNFKLDSPLYGTKIWGLCSQNKIEESEAVMREMK 243
Query: 354 GKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFE 413
GL N Y SL+ V ++ A+ L ++M+ + Y VLI LC+ G +
Sbjct: 244 DHGLTANSYIYTSLMDAYFKVGKITEAVNLLQEMQKFGIETTAVTYGVLIDGLCKKGLVQ 423
Query: 414 KGRELWDEATSMG 426
+ +D T G
Sbjct: 424 QAVSYFDCMTKTG 462
Score = 30.0 bits (66), Expect = 2.1
Identities = 18/85 (21%), Positives = 37/85 (43%)
Frame = +1
Query: 344 KGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLI 403
+ +E+ ++ G+ N + Y SLI + + A+++ ++M + P+Y I
Sbjct: 4 EAEELFRALLEAGVALNLQIYTSLIHGYMKAKMMEKAMDILKEMNKKNFKLDSPLYGTKI 183
Query: 404 PKLCRGGDFEKGRELWDEATSMGIT 428
LC E+ + E G+T
Sbjct: 184 WGLCSQNKIEESEAVMREMKDHGLT 258
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,321,695
Number of Sequences: 36976
Number of extensions: 247675
Number of successful extensions: 1679
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 1525
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1626
length of query: 495
length of database: 9,014,727
effective HSP length: 100
effective length of query: 395
effective length of database: 5,317,127
effective search space: 2100265165
effective search space used: 2100265165
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0093b.5


