

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093a.1
(268 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
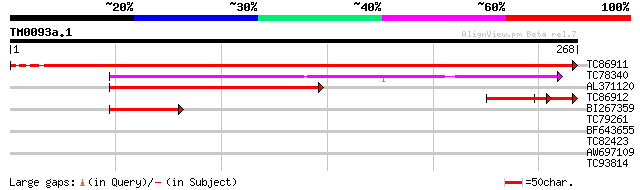
TC86911 homologue to GP|20339362|gb|AAM19354.1 ribulose-5-phosph... 488 e-138
TC78340 similar to GP|6007803|gb|AAF01048.1| D-ribulose-5-phosph... 154 4e-38
AL371120 similar to GP|6007803|gb| D-ribulose-5-phosphate 3-epim... 86 1e-17
TC86912 homologue to GP|20339362|gb|AAM19354.1 ribulose-5-phosph... 55 3e-12
BI267359 similar to PIR|G96658|G966 probable D-ribulose-5-phosph... 40 7e-04
TC79261 similar to PIR|T49047|T49047 quinone reductase-like prot... 32 0.34
BF643655 similar to GP|6056190|gb| putative glycerol-3-phosphate... 29 2.2
TC82423 similar to PIR|B86314|B86314 hypothetical protein AAF972... 29 2.2
AW697109 similar to GP|10177340|dbj transporter-like protein {Ar... 28 4.8
TC93814 similar to PIR|B86365|B86365 probable sulphate transport... 27 8.3
>TC86911 homologue to GP|20339362|gb|AAM19354.1
ribulose-5-phosphate-3-epimerase {Pisum sativum},
complete
Length = 1233
Score = 488 bits (1255), Expect = e-138
Identities = 252/268 (94%), Positives = 259/268 (96%)
Frame = +1
Query: 1 INGPSSLHSKTAIFNNHPRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKL 60
ING SLH K ++F HPRSLTFSRRKIST+VKASSRVDKFSKSDIIVSPSILSANFAKL
Sbjct: 88 ING-LSLH-KNSLF--HPRSLTFSRRKISTVVKASSRVDKFSKSDIIVSPSILSANFAKL 255
Query: 61 GEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVP 120
GEQVKAVELAGCDWIHVDVMDGRFVPNITIGPL+VDALRPVTDLPLDVHLMIVEP+QRVP
Sbjct: 256 GEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLVVDALRPVTDLPLDVHLMIVEPDQRVP 435
Query: 121 DFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVL 180
DFIKAGADIVSIHCEQSSTIHLHR +NQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVL
Sbjct: 436 DFIKAGADIVSIHCEQSSTIHLHRAINQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVL 615
Query: 181 IMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALV 240
IMSVNPGFGGQSFIESQVKKISDLRRLC EKGVNPWIEVDGGVTPANAYKVIEAGANALV
Sbjct: 616 IMSVNPGFGGQSFIESQVKKISDLRRLCVEKGVNPWIEVDGGVTPANAYKVIEAGANALV 795
Query: 241 AGSAVFGAKDYAEAIKGIKTSKRPEPVA 268
AGSAVFGAKDYAEAIKGIK SKRPEPV+
Sbjct: 796 AGSAVFGAKDYAEAIKGIKASKRPEPVS 879
>TC78340 similar to GP|6007803|gb|AAF01048.1| D-ribulose-5-phosphate
3-epimerase {Oryza sativa}, partial (95%)
Length = 1138
Score = 154 bits (388), Expect = 4e-38
Identities = 85/217 (39%), Positives = 130/217 (59%), Gaps = 3/217 (1%)
Frame = +3
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLD 107
++PS+LS++FA L + + G DW+H+D+MDG FVPN+T+G I+++LR T+ LD
Sbjct: 156 IAPSMLSSDFANLASEAHRMINYGADWLHMDIMDGHFVPNLTMGAPIIESLRKHTEAYLD 335
Query: 108 VHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLS 167
HLM+ P V KAGA + H E S + + +KS G + GV + PGT +
Sbjct: 336 CHLMVTNPLDYVEPLGKAGASGFTFHIETSKD-NWKELIQNIKSHGMRPGVSIKPGTSVE 512
Query: 168 AIEYVLDV---VDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVT 224
+ +++ V++VL+M+V PGFGGQ F+ + K+ LR+ K + IEVDGG+
Sbjct: 513 EVYPLVEAENPVEMVLVMTVEPGFGGQKFMPEMMDKVRILRK----KYPSLDIEVDGGLG 680
Query: 225 PANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTS 261
P+ AGAN +VAGS+VFGA + A I +++S
Sbjct: 681 PSTIDVAASAGANCIVAGSSVFGAPEPAHVISLLRSS 791
>AL371120 similar to GP|6007803|gb| D-ribulose-5-phosphate 3-epimerase {Oryza
sativa}, partial (56%)
Length = 448
Score = 85.9 bits (211), Expect = 1e-17
Identities = 41/101 (40%), Positives = 62/101 (60%)
Frame = +1
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLD 107
++PS+LS++FA L + + G DW+H+D+MDG FVPN+T+G I+++LR T+ LD
Sbjct: 46 IAPSMLSSDFANLASEAHRMINYGADWLHMDIMDGHFVPNLTMGAPIIESLRKHTEAYLD 225
Query: 108 VHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQ 148
HLM+ P V KAGA + H E S I + R + +
Sbjct: 226 CHLMVTNPLDYVEPLGKAGASGFTFHIETSKVIFVGRQLER 348
>TC86912 homologue to GP|20339362|gb|AAM19354.1
ribulose-5-phosphate-3-epimerase {Pisum sativum},
partial (15%)
Length = 474
Score = 55.5 bits (132), Expect(2) = 3e-12
Identities = 28/31 (90%), Positives = 29/31 (93%)
Frame = +2
Query: 226 ANAYKVIEAGANALVAGSAVFGAKDYAEAIK 256
ANAYKVIEAGANALVAGSAVFGAKDYA + K
Sbjct: 11 ANAYKVIEAGANALVAGSAVFGAKDYA*SYK 103
Score = 32.7 bits (73), Expect(2) = 3e-12
Identities = 15/20 (75%), Positives = 16/20 (80%)
Frame = +3
Query: 249 KDYAEAIKGIKTSKRPEPVA 268
K EAIKGIK SKRPEPV+
Sbjct: 81 KTMLEAIKGIKASKRPEPVS 140
>BI267359 similar to PIR|G96658|G966 probable D-ribulose-5-phosphate F9N12.9
[imported] - Arabidopsis thaliana, partial (17%)
Length = 415
Score = 40.4 bits (93), Expect = 7e-04
Identities = 15/35 (42%), Positives = 25/35 (70%)
Frame = +1
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDG 82
++PS+LS++FA L + + G DW+H+D+MDG
Sbjct: 178 IAPSMLSSDFANLASEAHRMINYGADWLHMDIMDG 282
>TC79261 similar to PIR|T49047|T49047 quinone reductase-like protein -
Arabidopsis thaliana, partial (95%)
Length = 1239
Score = 31.6 bits (70), Expect = 0.34
Identities = 59/230 (25%), Positives = 93/230 (39%), Gaps = 27/230 (11%)
Frame = +3
Query: 42 SKSDIIVSPSILSANFA-------KLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLI 94
S + + V S NFA K E+ + G D+ + G V N +G
Sbjct: 132 SPTSVRVRIKATSLNFANYLQILGKYQEKPHLPFIPGSDFSGIVDSVGSNVTNFRVG--- 302
Query: 95 VDALRPVTDLPLDVHLMIVEPEQ--RVP---DFIKAGADIVSIHCEQSSTIHLHRTVNQV 149
D + L ++V+ Q RVP D + AGA V+ +H Q+
Sbjct: 303 -DPVCSFAALGSYAQYLVVDQTQLFRVPEGCDLVAAGALAVAFGTSHVGLVHRA----QL 467
Query: 150 KS------LGAKAGVVLNP-------GTPLSAIEYVLDVVDLVLIMSVNP--GFGGQSFI 194
KS LGA GV L G + A+ + V L+ M V+ G ++
Sbjct: 468 KSGQVLLVLGAAGGVGLAAVQIGKACGAIVIAVARGAEKVQLLKSMGVDHVVDLGNENVT 647
Query: 195 ESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSA 244
ES VK+ ++RL KG++ + GG + ++++ GAN L+ G A
Sbjct: 648 ES-VKEFLKVKRL---KGIDVLYDPVGGKLMKESLRLLKWGANILIIGFA 785
>BF643655 similar to GP|6056190|gb| putative glycerol-3-phosphate
dehydrogenase {Arabidopsis thaliana}, partial (27%)
Length = 587
Score = 28.9 bits (63), Expect = 2.2
Identities = 18/46 (39%), Positives = 24/46 (52%)
Frame = +1
Query: 18 PRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQ 63
PRS +RR ST SSR DKFS + P + + F+ L E+
Sbjct: 31 PRSQLSNRR--STTQTPSSRPDKFSSRHSLAPPPLTRSTFS*LAEE 162
>TC82423 similar to PIR|B86314|B86314 hypothetical protein AAF97269.1
[imported] - Arabidopsis thaliana, partial (51%)
Length = 748
Score = 28.9 bits (63), Expect = 2.2
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Frame = +2
Query: 131 SIHCEQSSTIH-LHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIM 182
+ H E S + L R ++ K GAK VV G+PL +V D+ D V+ M
Sbjct: 95 NFHPENSHVLPALMRRFHEAKVNGAKEVVVWGTGSPLREFLHVDDLADAVVFM 253
>AW697109 similar to GP|10177340|dbj transporter-like protein {Arabidopsis
thaliana}, partial (17%)
Length = 651
Score = 27.7 bits (60), Expect = 4.8
Identities = 16/54 (29%), Positives = 31/54 (56%), Gaps = 6/54 (11%)
Frame = -1
Query: 6 SLHSKTAIFNNHPRSLTFSRRKISTIVKASSRVDK------FSKSDIIVSPSIL 53
++ KTA+ N+HP+ +R K V+ +R + F+++D+++SP IL
Sbjct: 618 NIQEKTAVVNSHPK----TRAKAK*CVEDDNRDNP*NCHRFFTRNDLLLSPMIL 469
>TC93814 similar to PIR|B86365|B86365 probable sulphate transporter protein
[imported] - Arabidopsis thaliana, partial (30%)
Length = 671
Score = 26.9 bits (58), Expect = 8.3
Identities = 9/32 (28%), Positives = 20/32 (62%)
Frame = +2
Query: 83 RFVPNITIGPLIVDALRPVTDLPLDVHLMIVE 114
++ PN+ +G +IV A+ + D+P H+ ++
Sbjct: 398 QYTPNVVLGAIIVTAVIGLIDIPAACHIWKID 493
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,816,211
Number of Sequences: 36976
Number of extensions: 78609
Number of successful extensions: 411
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 407
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 409
length of query: 268
length of database: 9,014,727
effective HSP length: 94
effective length of query: 174
effective length of database: 5,538,983
effective search space: 963783042
effective search space used: 963783042
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0093a.1


