

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
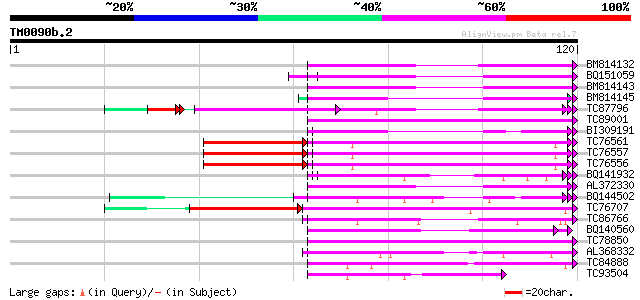
Query= TM0090b.2
(120 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 59 3e-10
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 56 3e-09
BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f... 56 3e-09
BM814145 homologue to GP|21322711|em pherophorin-dz1 protein {Vo... 54 1e-08
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 54 1e-08
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 52 4e-08
BI309191 52 7e-08
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 49 9e-08
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 49 9e-08
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 49 9e-08
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 50 2e-07
AL372330 similar to GP|21322752|dbj cold shock protein-1 {Tritic... 48 8e-07
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 47 1e-06
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 42 2e-06
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 47 2e-06
BQ140560 weakly similar to GP|15128223|db hypothetical protein~s... 45 5e-06
TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like ... 44 1e-05
AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein... 43 2e-05
TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 40 2e-04
TC93504 weakly similar to PIR|S46286|S46286 RNA-binding protein ... 39 6e-04
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 59.3 bits (142), Expect = 3e-10
Identities = 29/57 (50%), Positives = 30/57 (51%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG R G GGG G+ GGG GGGGGGGG
Sbjct: 10 GGGGGGGGGGGGGGGGGGRGGGG-------------GGGGGGAGGGGGGGGGGGGGG 141
Score = 58.5 bits (140), Expect = 6e-10
Identities = 29/57 (50%), Positives = 29/57 (50%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GR GGG G GGG GGGGGGGG
Sbjct: 29 GGGGGGGGGGGGGGGGGGGGGGGR------------GGGGGGGGGGGGGGGGGGGGG 163
Score = 58.2 bits (139), Expect = 7e-10
Identities = 29/57 (50%), Positives = 29/57 (50%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G R GGG G GGG GGGGGGGG
Sbjct: 26 GGGGGGGGGGGGGGGGGGGGGGGGR------------GGGGGGGGGGGGGGGGGGGG 160
Score = 55.5 bits (132), Expect = 5e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 1 GGGGGGGGGGGGGGGGGGGGGRG------------GGGGGGGGGAGGGGGGGGGGGG 135
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 113 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 241
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 37 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 165
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 35 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 163
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 76 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 204
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 79 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 207
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 82 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 210
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 93 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 221
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 41 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 169
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 150 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 278
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 154 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 282
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 161 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 289
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 163 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 291
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 169 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 297
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 174 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 302
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 175 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 303
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 178 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 306
Score = 50.8 bits (120), Expect = 1e-07
Identities = 26/55 (47%), Positives = 26/55 (47%)
Frame = +2
Query: 66 GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G N GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 23 GENPGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 145
Score = 48.5 bits (114), Expect = 6e-07
Identities = 25/61 (40%), Positives = 27/61 (43%)
Frame = +2
Query: 60 VVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
+ + G GGG GG GGG G GGG G GGG GGGGGGG
Sbjct: 11 IPLSGENPGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGG 148
Query: 120 G 120
G
Sbjct: 149 G 151
>BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (4%)
Length = 132
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 4 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 132
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 2 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 130
>BM814145 homologue to GP|21322711|em pherophorin-dz1 protein {Volvox carteri
f. nagariensis}, partial (3%)
Length = 111
Score = 53.9 bits (128), Expect = 1e-08
Identities = 27/57 (47%), Positives = 27/57 (47%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 1 GGGGGGGGGGGGGGGGG--------------------GGGGGGGGGGGGGGGGGGGG 111
Score = 41.2 bits (95), Expect = 9e-05
Identities = 23/58 (39%), Positives = 23/58 (39%)
Frame = +2
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
V GGGG GGG GG GGG GGG GGGGGGG
Sbjct: 20 VGGGGGGGGGGGGGGGGGG----------------------------GGGGGGGGGGG 109
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 53.9 bits (128), Expect = 1e-08
Identities = 27/57 (47%), Positives = 30/57 (52%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG +GG +G GGG+G GGGS GGGGGGG
Sbjct: 25 GGGGGSGGGGGGGAGGAHGVGYG-------------SGGGTGGGYGGGSPGGGGGGG 156
Score = 43.9 bits (102), Expect = 1e-05
Identities = 34/100 (34%), Positives = 35/100 (35%)
Frame = +1
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG SGGGG GGG G G SGGG
Sbjct: 28 GGGGSGGGG----------------------------------GGGAGGAHGVGYGSGGG 105
Query: 81 ARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+G GGG G GGGSGGGGGGGG
Sbjct: 106 TGGGYGGG----------SPGGGGG---GGGSGGGGGGGG 186
Score = 42.7 bits (99), Expect(3) = 1e-07
Identities = 24/56 (42%), Positives = 24/56 (42%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GGG GG SGGG GG G GGG GGG GGG
Sbjct: 121 GGGSPGGGGGGGGSGGGGGG-----------------GGAHGGAYGGGIGGGEGGG 237
Score = 42.4 bits (98), Expect(2) = 4e-07
Identities = 23/55 (41%), Positives = 23/55 (41%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
GGGG GG GG GGGA GG G GGG GGG GG
Sbjct: 136 GGGGGGGGSGGGGGGGGAH------------------GGAYGGGIGGGEGGGHGG 246
Score = 42.4 bits (98), Expect = 4e-05
Identities = 26/66 (39%), Positives = 30/66 (45%), Gaps = 9/66 (13%)
Frame = +1
Query: 64 GGGGNDGGGSGGS---------SGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG 114
GGGG+ GGG GG+ SGGG +G GGG G GG+ G
Sbjct: 28 GGGGSGGGGGGGAGGAHGVGYGSGGGTGGGYGGGSPGGGGGGGGSGGGGGGGGAHGGAYG 207
Query: 115 GGGGGG 120
GG GGG
Sbjct: 208 GGIGGG 225
Score = 35.0 bits (79), Expect = 0.007
Identities = 14/22 (63%), Positives = 14/22 (63%)
Frame = +1
Query: 99 DDGGGSGSDDGGGSGGGGGGGG 120
D GGG G G G GGGGG GG
Sbjct: 7 DHGGGYGGGGGSGGGGGGGAGG 72
Score = 30.8 bits (68), Expect = 0.13
Identities = 12/16 (75%), Positives = 12/16 (75%)
Frame = +1
Query: 105 GSDDGGGSGGGGGGGG 120
G D GGG GGGGG GG
Sbjct: 1 GRDHGGGYGGGGGSGG 48
Score = 26.2 bits (56), Expect(2) = 4e-07
Identities = 6/8 (75%), Positives = 6/8 (75%)
Frame = +3
Query: 30 WCWWRRWR 37
WCWW WR
Sbjct: 60 WCWWSAWR 83
Score = 25.8 bits (55), Expect = 4.1
Identities = 10/18 (55%), Positives = 12/18 (66%), Gaps = 4/18 (22%)
Frame = +3
Query: 85 WGRRRQW---WR-WRQWC 98
W R R+W WR WR+WC
Sbjct: 12 WRRIRRWRWKWRWWRRWC 65
Score = 24.3 bits (51), Expect(3) = 1e-07
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = +3
Query: 30 WCWWRRW 36
W WWRRW
Sbjct: 42 WRWWRRW 62
Score = 22.3 bits (46), Expect(3) = 1e-07
Identities = 14/31 (45%), Positives = 17/31 (54%)
Frame = +2
Query: 40 VVVVAVTVVVIVVEAVVVEAVVVDGGGGNDG 70
VV V V + ++ VVVE VVD G G G
Sbjct: 47 VVEEVVLVERMALDTVVVEEPVVDMGVGRLG 139
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 52.4 bits (124), Expect = 4e-08
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG G GGG G RR R G G G D G G GGGGG GG
Sbjct: 315 GGGGGYGGGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGGGGDRGYGGGGGGGYGG 485
Score = 32.0 bits (71), Expect = 0.057
Identities = 13/19 (68%), Positives = 13/19 (68%)
Frame = +3
Query: 101 GGGSGSDDGGGSGGGGGGG 119
GGG G GGG GGGGGG
Sbjct: 312 GGGGGGYGGGGGYGGGGGG 368
Score = 32.0 bits (71), Expect = 0.057
Identities = 13/19 (68%), Positives = 13/19 (68%)
Frame = +3
Query: 102 GGSGSDDGGGSGGGGGGGG 120
GG G GGG G GGGGGG
Sbjct: 312 GGGGGGYGGGGGYGGGGGG 368
Score = 31.2 bits (69), Expect = 0.097
Identities = 13/20 (65%), Positives = 13/20 (65%)
Frame = +3
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
GGG G GG GGGG GGG
Sbjct: 300 GGGRGGGGGGYGGGGGYGGG 359
Score = 31.2 bits (69), Expect = 0.097
Identities = 13/20 (65%), Positives = 13/20 (65%)
Frame = +3
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
GGG GGG GGGGG GG
Sbjct: 297 GGGGRGGGGGGYGGGGGYGG 356
Score = 30.0 bits (66), Expect = 0.22
Identities = 12/19 (63%), Positives = 12/19 (63%)
Frame = +3
Query: 102 GGSGSDDGGGSGGGGGGGG 120
G G GGG GG GGGGG
Sbjct: 291 GSGGGGRGGGGGGYGGGGG 347
Score = 30.0 bits (66), Expect = 0.22
Identities = 12/19 (63%), Positives = 12/19 (63%)
Frame = +3
Query: 101 GGGSGSDDGGGSGGGGGGG 119
G G G GGG G GGGGG
Sbjct: 291 GSGGGGRGGGGGGYGGGGG 347
Score = 25.8 bits (55), Expect = 4.1
Identities = 8/14 (57%), Positives = 8/14 (57%)
Frame = +2
Query: 84 WWGRRRQWWRWRQW 97
WW R R W RW W
Sbjct: 296 WWRR*RWWRRWLWW 337
Score = 25.0 bits (53), Expect = 6.9
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = +2
Query: 30 WCWWRRW 36
W WWRRW
Sbjct: 326 WLWWRRW 346
>BI309191
Length = 109
Score = 51.6 bits (122), Expect = 7e-08
Identities = 26/56 (46%), Positives = 26/56 (46%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGG GGG G GGG GGGGGGG
Sbjct: 1 GGGGGGGGGGGGGGGGG--------------------GGGGGGGGGGGGGGGGGGG 108
Score = 51.6 bits (122), Expect = 7e-08
Identities = 26/56 (46%), Positives = 26/56 (46%)
Frame = +1
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 1 GGGGGGGGGGGGGGGG--------------------GGGGGGGGGGGGGGGGGGGG 108
Score = 49.7 bits (117), Expect = 3e-07
Identities = 27/57 (47%), Positives = 27/57 (47%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 3 GGGGGGGGGGGGGGGGGG-------------------GGGGG---GGGGGGGGGGGG 107
Score = 49.3 bits (116), Expect = 3e-07
Identities = 27/57 (47%), Positives = 27/57 (47%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 8 GGGGGGGGGGGGGGGGG--------------------GGGGG---GGGGGGGGGGGG 109
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 48.9 bits (115), Expect = 4e-07
Identities = 26/59 (44%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Frame = +1
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG---GGGGGG 120
GGG GG GG GGG + G RR + + GGG + GG GG GGGGGG
Sbjct: 601 GGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGG 777
Score = 48.5 bits (114), Expect(2) = 9e-08
Identities = 28/61 (45%), Positives = 29/61 (46%), Gaps = 5/61 (8%)
Frame = +1
Query: 64 GGGGNDGG-----GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
GGGG GG G GG GGG +G RR GGG G GGG GG GGG
Sbjct: 643 GGGGGYGGERRGYGGGGGYGGGGGGGYGERR-----------GGGGGYSRGGGGGGYGGG 789
Query: 119 G 119
G
Sbjct: 790 G 792
Score = 26.6 bits (57), Expect(2) = 0.23
Identities = 10/19 (52%), Positives = 10/19 (52%), Gaps = 3/19 (15%)
Frame = +3
Query: 82 RAWWGRRRQWW---RWRQW 97
R WW R WW RWR W
Sbjct: 759 RRWWWRIWWWWIQQRWR*W 815
Score = 25.4 bits (54), Expect = 5.3
Identities = 12/17 (70%), Positives = 12/17 (70%)
Frame = +1
Query: 103 GSGSDDGGGSGGGGGGG 119
GSG GGG GG GGGG
Sbjct: 595 GSG---GGGGGGRGGGG 636
Score = 25.0 bits (53), Expect = 6.9
Identities = 10/15 (66%), Positives = 10/15 (66%)
Frame = +1
Query: 106 SDDGGGSGGGGGGGG 120
S GG GGGG GGG
Sbjct: 589 SRGSGGGGGGGRGGG 633
Score = 22.3 bits (46), Expect(2) = 9e-08
Identities = 13/22 (59%), Positives = 14/22 (63%)
Frame = +2
Query: 42 VVAVTVVVIVVEAVVVEAVVVD 63
VVAV V V AVV+E V VD
Sbjct: 593 VVAVEVEEEAVVAVVMEVVAVD 658
Score = 21.9 bits (45), Expect(2) = 0.23
Identities = 13/24 (54%), Positives = 14/24 (58%)
Frame = +2
Query: 40 VVVVAVTVVVIVVEAVVVEAVVVD 63
V VV V V +VE VVV VVD
Sbjct: 626 VAVVMEVVAVDMVENVVVTVEVVD 697
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 48.9 bits (115), Expect = 4e-07
Identities = 26/59 (44%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Frame = +1
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG---GGGGGG 120
GGG GG GG GGG + G RR + + GGG + GG GG GGGGGG
Sbjct: 505 GGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGG 681
Score = 48.5 bits (114), Expect(2) = 9e-08
Identities = 28/61 (45%), Positives = 29/61 (46%), Gaps = 5/61 (8%)
Frame = +1
Query: 64 GGGGNDGG-----GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
GGGG GG G GG GGG +G RR GGG G GGG GG GGG
Sbjct: 547 GGGGGYGGERRGYGGGGGYGGGGGGGYGERR-----------GGGGGYSRGGGGGGYGGG 693
Query: 119 G 119
G
Sbjct: 694 G 696
Score = 26.6 bits (57), Expect(2) = 0.23
Identities = 10/19 (52%), Positives = 10/19 (52%), Gaps = 3/19 (15%)
Frame = +3
Query: 82 RAWWGRRRQWW---RWRQW 97
R WW R WW RWR W
Sbjct: 663 RRWWWRIWWWWIQQRWR*W 719
Score = 25.4 bits (54), Expect = 5.3
Identities = 12/17 (70%), Positives = 12/17 (70%)
Frame = +1
Query: 103 GSGSDDGGGSGGGGGGG 119
GSG GGG GG GGGG
Sbjct: 499 GSG---GGGGGGRGGGG 540
Score = 25.0 bits (53), Expect = 6.9
Identities = 10/15 (66%), Positives = 10/15 (66%)
Frame = +1
Query: 106 SDDGGGSGGGGGGGG 120
S GG GGGG GGG
Sbjct: 493 SRGSGGGGGGGRGGG 537
Score = 22.3 bits (46), Expect(2) = 9e-08
Identities = 13/22 (59%), Positives = 14/22 (63%)
Frame = +2
Query: 42 VVAVTVVVIVVEAVVVEAVVVD 63
VVAV V V AVV+E V VD
Sbjct: 497 VVAVEVEEEAVVAVVMEVVAVD 562
Score = 21.9 bits (45), Expect(2) = 0.23
Identities = 13/24 (54%), Positives = 14/24 (58%)
Frame = +2
Query: 40 VVVVAVTVVVIVVEAVVVEAVVVD 63
V VV V V +VE VVV VVD
Sbjct: 530 VAVVMEVVAVDMVENVVVTVEVVD 601
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 48.9 bits (115), Expect = 4e-07
Identities = 26/59 (44%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Frame = +2
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG---GGGGGG 120
GGG GG GG GGG + G RR + + GGG + GG GG GGGGGG
Sbjct: 347 GGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGG 523
Score = 48.5 bits (114), Expect(2) = 9e-08
Identities = 28/61 (45%), Positives = 29/61 (46%), Gaps = 5/61 (8%)
Frame = +2
Query: 64 GGGGNDGG-----GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
GGGG GG G GG GGG +G RR GGG G GGG GG GGG
Sbjct: 389 GGGGGYGGERRGYGGGGGYGGGGGGGYGERR-----------GGGGGYSRGGGGGGYGGG 535
Query: 119 G 119
G
Sbjct: 536 G 538
Score = 26.6 bits (57), Expect(2) = 0.24
Identities = 10/19 (52%), Positives = 10/19 (52%), Gaps = 3/19 (15%)
Frame = +1
Query: 82 RAWWGRRRQWW---RWRQW 97
R WW R WW RWR W
Sbjct: 505 RRWWWRIWWWWIQQRWR*W 561
Score = 25.4 bits (54), Expect = 5.3
Identities = 12/17 (70%), Positives = 12/17 (70%)
Frame = +2
Query: 103 GSGSDDGGGSGGGGGGG 119
GSG GGG GG GGGG
Sbjct: 341 GSG---GGGGGGRGGGG 382
Score = 25.0 bits (53), Expect = 6.9
Identities = 10/15 (66%), Positives = 10/15 (66%)
Frame = +2
Query: 106 SDDGGGSGGGGGGGG 120
S GG GGGG GGG
Sbjct: 335 SRGSGGGGGGGRGGG 379
Score = 22.3 bits (46), Expect(2) = 9e-08
Identities = 13/22 (59%), Positives = 14/22 (63%)
Frame = +3
Query: 42 VVAVTVVVIVVEAVVVEAVVVD 63
VVAV V V AVV+E V VD
Sbjct: 339 VVAVEVEEEAVVAVVMEVVAVD 404
Score = 21.9 bits (45), Expect(2) = 0.24
Identities = 13/24 (54%), Positives = 14/24 (58%)
Frame = +3
Query: 40 VVVVAVTVVVIVVEAVVVEAVVVD 63
V VV V V +VE VVV VVD
Sbjct: 372 VAVVMEVVAVDMVENVVVTVEVVD 443
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 50.1 bits (118), Expect = 2e-07
Identities = 28/61 (45%), Positives = 31/61 (49%), Gaps = 6/61 (9%)
Frame = +2
Query: 66 GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDD------GGGSGGGGGGG 119
GG GGG GG++GGG W R W C GGG+ DD GG GG GGGG
Sbjct: 278 GGGVGGGGGGTAGGGIPQWVQRAEGEWN----CRCGGGARVDDRGRPGRGGEVGGWGGGG 445
Query: 120 G 120
G
Sbjct: 446 G 448
Score = 49.3 bits (116), Expect = 3e-07
Identities = 29/67 (43%), Positives = 31/67 (45%), Gaps = 12/67 (17%)
Frame = +1
Query: 65 GGGNDGGGSGGSSGGGAR---------AWWGRRRQWWRWRQWCDDGGGSGSDDGGGS--- 112
G G +GGG GG GGG AW GR W W GGG G D GGG+
Sbjct: 160 GRGGEGGGWGGGGGGGGLVRGRRKEEGAWGGRDESWGGG--WRGSGGGGGGDGGGGNPAM 333
Query: 113 GGGGGGG 119
G GGGG
Sbjct: 334 GTKGGGG 354
Score = 46.6 bits (109), Expect = 2e-06
Identities = 26/58 (44%), Positives = 30/58 (50%), Gaps = 3/58 (5%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGG---SGSDDGGGSGGGGGG 118
GGGG GGG G G G G+R+ GGG +G +DGGG GGGGGG
Sbjct: 161 GGGGRGGGGGGEGEGEGWLGGGGKRK---------GRGGGETRAGGEDGGGVGGGGGG 307
Score = 37.7 bits (86), Expect = 0.001
Identities = 25/57 (43%), Positives = 26/57 (44%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG G G RAW G R W GG+G GGS GGGG G
Sbjct: 412 GGGGRRVGGRRG--GERRRAWVGGRTGW----------GGAGRGVAGGSVQGGGGLG 546
Score = 36.2 bits (82), Expect = 0.003
Identities = 16/25 (64%), Positives = 17/25 (68%)
Frame = +2
Query: 96 QWCDDGGGSGSDDGGGSGGGGGGGG 120
+WC D GG G GGG GGGGGG G
Sbjct: 131 RWCLDWGGCGG--GGGRGGGGGGEG 199
Score = 33.9 bits (76), Expect = 0.015
Identities = 27/84 (32%), Positives = 29/84 (34%), Gaps = 27/84 (32%)
Frame = +1
Query: 64 GGGGNDGGGSG---------------------------GSSGGGARAWWGRRRQWWRWRQ 96
GGGG DGGG G+ GGG R G RR R R
Sbjct: 292 GGGGGDGGGGNPAMGTKGGGGVELPMRRGGEGR*PWATGAGGGGRRV--GGRRGGERRRA 465
Query: 97 WCDDGGGSGSDDGGGSGGGGGGGG 120
W G G G +GG GGG
Sbjct: 466 WVGGRTGWGGAGRGVAGGSVQGGG 537
Score = 33.1 bits (74), Expect = 0.025
Identities = 23/57 (40%), Positives = 26/57 (45%), Gaps = 2/57 (3%)
Frame = +3
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRR--QWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG +GG + GGGA G R+ WW GSG G GG GGGG
Sbjct: 429 GGGAEGGRAKARVGGGADGMGGGRQGCGWW---------VGSGWRGIGMLGG*GGGG 572
Score = 32.7 bits (73), Expect = 0.033
Identities = 26/69 (37%), Positives = 28/69 (39%), Gaps = 9/69 (13%)
Frame = +3
Query: 60 VVVDGGGGNDGGGSGGSSGGG---------ARAWWGRRRQWWRWRQWCDDGGGSGSDDGG 110
VVV GGG GGG GG G G R G RR+ GG + G
Sbjct: 150 VVVGAGGG--GGGVGGGRGRGRAG*GEEERGRGVGGERREL----------GGRMEGEWG 293
Query: 111 GSGGGGGGG 119
G GGG GG
Sbjct: 294 GGGGGRRGG 320
Score = 31.6 bits (70), Expect = 0.074
Identities = 25/61 (40%), Positives = 29/61 (46%), Gaps = 6/61 (9%)
Frame = +3
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQ---WCDDGGGSGSDDGGGSGGGG---GG 118
GG +G GG GGG R G R ++ R+ D GG GS G GGGG GG
Sbjct: 267 GGRMEGEWGGG--GGGRRG--GESRNGYKGRRGSGTADAAGGRGSMTVGDRGGGGRSEGG 434
Query: 119 G 119
G
Sbjct: 435 G 437
Score = 31.2 bits (69), Expect = 0.097
Identities = 25/70 (35%), Positives = 26/70 (36%), Gaps = 13/70 (18%)
Frame = +2
Query: 64 GGGGNDGG-GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGG------------SGSDDGG 110
G GG GG G GG GAR W G R DGGG +G D G
Sbjct: 407 GRGGEVGGWGGGGGESEGARGWGGGR-----------DGGGQAGVWLVGRFRVAGDWDAG 553
Query: 111 GSGGGGGGGG 120
G G GG
Sbjct: 554 GLGWWWMAGG 583
Score = 27.3 bits (59), Expect = 1.4
Identities = 13/28 (46%), Positives = 14/28 (49%)
Frame = +1
Query: 89 RQWWRWRQWCDDGGGSGSDDGGGSGGGG 116
R W W + GG G GGG GGGG
Sbjct: 142 RLGWLWGR-----GGEGGGWGGGGGGGG 210
>AL372330 similar to GP|21322752|dbj cold shock protein-1 {Triticum
aestivum}, partial (33%)
Length = 435
Score = 48.1 bits (113), Expect = 8e-07
Identities = 26/57 (45%), Positives = 28/57 (48%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG+ GG GG GGGA +G GGG G GGG GGGG GGG
Sbjct: 146 GYGGSSGGYGGGGYGGGASGGYG--------------GGGYGGGSGGGYGGGGYGGG 274
Score = 45.4 bits (106), Expect = 5e-06
Identities = 26/56 (46%), Positives = 26/56 (46%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GG SGG GGG G G G G GGG GGGGGGG
Sbjct: 173 GGGGYGGGASGGYGGGGYGGGSG-------------GGYGGGGYGGGGYGGGGGGG 301
Score = 35.4 bits (80), Expect = 0.005
Identities = 24/69 (34%), Positives = 27/69 (38%), Gaps = 13/69 (18%)
Frame = +2
Query: 64 GGGGNDGGGSGGSS-------------GGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGG 110
G GG+ GG GGSS GGG+ +G GG G GG
Sbjct: 26 GYGGSSSGGYGGSSSSSYPASSSGGGYGGGSSGGYG------------GSSGGYGGSSGG 169
Query: 111 GSGGGGGGG 119
GGG GGG
Sbjct: 170 YGGGGYGGG 196
Score = 34.3 bits (77), Expect = 0.011
Identities = 13/26 (50%), Positives = 14/26 (53%), Gaps = 6/26 (23%)
Frame = +1
Query: 82 RAWWGR------RRQWWRWRQWCDDG 101
R WW R RR WWRW +W DG
Sbjct: 244 RIWWRRLWWRWLRRWWWRWLRWRQDG 321
Score = 33.5 bits (75), Expect = 0.019
Identities = 19/54 (35%), Positives = 23/54 (42%)
Frame = +2
Query: 67 GNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG S G GG + + + + G G S GGS GG GGGG
Sbjct: 23 GGYGGSSSGGYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGGYGGSSGGYGGGG 184
Score = 27.3 bits (59), Expect = 1.4
Identities = 13/30 (43%), Positives = 13/30 (43%)
Frame = +2
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRW 94
GGG GGG GG GG G Q W
Sbjct: 266 GGGGYGGGGGGGYGGDKMGALGANLQKQDW 355
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 47.4 bits (111), Expect = 1e-06
Identities = 32/99 (32%), Positives = 34/99 (34%)
Frame = +3
Query: 22 GGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGGA 81
GG+ GG GWC W GGGG GGG G GGG
Sbjct: 72 GGEVGGSGWCVW------------------------------GGGG--GGGRCGGGGGG- 152
Query: 82 RAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G + G GGGGGGGG
Sbjct: 153 -------------------GGGRGGAEAAGPGGGGGGGG 212
Score = 44.3 bits (103), Expect = 1e-05
Identities = 25/62 (40%), Positives = 27/62 (43%), Gaps = 2/62 (3%)
Frame = +3
Query: 61 VVDGGGGNDGGG--SGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
+ +GGG GG GG GG WG GGG G GGG GGGGG
Sbjct: 27 IPEGGGARGAGGWRRGGEVGGSGWCVWG--------------GGGGGGRCGGGGGGGGGR 164
Query: 119 GG 120
GG
Sbjct: 165 GG 170
Score = 41.6 bits (96), Expect = 7e-05
Identities = 28/63 (44%), Positives = 29/63 (45%), Gaps = 7/63 (11%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGAR----AWWGRR---RQWWRWRQWCDDGGGSGSDDGGGSGGGG 116
GGG GG + G GGG R WGR R W GGG G GGG GG G
Sbjct: 38 GGGAGGGGLAAGGGGGGLRLVRVGGWGRGGPLRGGW--------GGGRGPR-GGGGGGPG 190
Query: 117 GGG 119
GGG
Sbjct: 191 GGG 199
Score = 38.5 bits (88), Expect = 6e-04
Identities = 22/58 (37%), Positives = 26/58 (43%), Gaps = 3/58 (5%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDD---GGGSGSDDGGGSGGGGGG 118
G G DGGG G G G R + R + + C G+ S GGG GG GGG
Sbjct: 514 GALGGDGGGGTGGGGRGGRGYVATRTRGTSAGRGCPPIFLARGTRSHSGGGGGGRGGG 687
Score = 37.4 bits (85), Expect = 0.001
Identities = 20/46 (43%), Positives = 22/46 (47%)
Frame = +1
Query: 74 GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GG GGGA + D GG G D GGG+GGGG GG
Sbjct: 475 GGGGGGGAAS---------------DCGGALGGDGGGGTGGGGRGG 567
Score = 37.0 bits (84), Expect = 0.002
Identities = 29/82 (35%), Positives = 31/82 (37%), Gaps = 24/82 (29%)
Frame = +1
Query: 62 VDGGGGNDGGGSGGSSGG-----------------GARAW-------WGRRRQWWRWRQW 97
V GGG G GSG GAR W GRR R +
Sbjct: 298 VGGGGRGRGRGSGPRCAALGEFPDLGPGTTATP*RGARGWPAGSGALGGRRGARGRAPRG 477
Query: 98 CDDGGGSGSDDGGGSGGGGGGG 119
GGG+ SD GG GG GGGG
Sbjct: 478 GGGGGGAASDCGGALGGDGGGG 543
Score = 37.0 bits (84), Expect = 0.002
Identities = 27/58 (46%), Positives = 28/58 (47%), Gaps = 3/58 (5%)
Frame = +1
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGG---GSGGGGGGG 119
GG GG + G GGGA A GRRR R R GG S GG G G G GGG
Sbjct: 106 GGVGAGGAAAGGVGGGAGAEGGRRR---RARGGGAVAGGMPSAAGGLLRGVGPGAGGG 270
Score = 36.6 bits (83), Expect = 0.002
Identities = 23/57 (40%), Positives = 25/57 (43%), Gaps = 5/57 (8%)
Frame = +2
Query: 65 GGGNDGGGSGGSSGGGAR-----AWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG 116
GGG GG GG +GGGAR WG G G+G GGG GGG
Sbjct: 446 GGGRAGGRRGGGAGGGARPRIVEGLWG--------------GTGAGGLAGGGEVGGG 574
Score = 36.6 bits (83), Expect = 0.002
Identities = 28/84 (33%), Positives = 31/84 (36%), Gaps = 28/84 (33%)
Frame = +3
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQ---WWR--WRQW--------------------CD 99
G G GGG S GGG WGR Q W R W
Sbjct: 243 GSGAGGGGGARSRGGGGPGGWGRAGQGTGLWTPVCRPWGVS*PWPGHHGDALTGGARVAG 422
Query: 100 DGGGSGSDDGG---GSGGGGGGGG 120
GG+G +GG G+ GGG GGG
Sbjct: 423 GVGGAGGPEGGARAGAAGGGRGGG 494
Score = 34.7 bits (78), Expect = 0.009
Identities = 26/66 (39%), Positives = 27/66 (40%), Gaps = 9/66 (13%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRR-------QWWRWRQWCDDGGG--SGSDDGGGSGG 114
GGG GG GG GGG R G R +W R R GGG G G G
Sbjct: 146 GGGRGPRGGGGGGPGGGGRWRGGCPRPQAAC*GEWGRGR-----GGGPVQGRRGAGWVGE 310
Query: 115 GGGGGG 120
GG G G
Sbjct: 311 GGAGDG 328
Score = 34.3 bits (77), Expect = 0.011
Identities = 25/64 (39%), Positives = 28/64 (43%), Gaps = 7/64 (10%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGG-------ARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG 116
GG G G GG GGG RA G Q+ W++ G GG GGGG
Sbjct: 524 GGTGAGGLAGGGEVGGGM*RLGREGRAPGGAAHQF-SWQE------GRVPIPGGEEGGGG 682
Query: 117 GGGG 120
GGGG
Sbjct: 683 GGGG 694
Score = 33.5 bits (75), Expect = 0.019
Identities = 21/56 (37%), Positives = 24/56 (42%)
Frame = +3
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG 117
V G GG +GG G++GGG GR WR GGG G D G G
Sbjct: 426 VGGAGGPEGGARAGAAGGGRGG--GRGLGLWR-----GSGGGRGRGDWRGGARWAG 572
Score = 32.3 bits (72), Expect = 0.043
Identities = 20/60 (33%), Positives = 24/60 (39%)
Frame = +1
Query: 60 VVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
++ GGG G G+GG GGG RW GG G+ G GGG G
Sbjct: 22 IIYQKGGGRGGRGAGG--GGG------------RWGAQAGACGGVGAGGAAAGGVGGGAG 159
Score = 31.6 bits (70), Expect = 0.074
Identities = 23/57 (40%), Positives = 27/57 (47%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G GG +GG GGGA R W GG+G+ GG +GGG GGG
Sbjct: 431 GRWGAGGGRAGGRRGGGAGGGARPRIVEGLW-------GGTGA--GGLAGGGEVGGG 574
Score = 31.2 bits (69), Expect = 0.097
Identities = 28/83 (33%), Positives = 29/83 (34%), Gaps = 28/83 (33%)
Frame = +3
Query: 64 GGGGNDGGGSG---GSSGGGARAWWGRRRQWWRWRQWCDD-------------------- 100
GGG G G G GS GG R W R RW C D
Sbjct: 474 GGGRGGGRGLGLWRGSGGGRGRGDW---RGGARWAGVCSDSDERDERRAGLPTNFLGKRD 644
Query: 101 -----GGGSGSDDGGGSGGGGGG 118
GG G+ GGG GGGGG
Sbjct: 645 AFPFRGGRRGA--GGGEEGGGGG 707
Score = 27.3 bits (59), Expect = 1.4
Identities = 16/40 (40%), Positives = 19/40 (47%)
Frame = +2
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGG 102
+GGGG GGG + G A RR + WR W GG
Sbjct: 668 EGGGGGGGGGWRRGAREGCCAALARRVRLGGWR-WAVRGG 784
Score = 26.6 bits (57), Expect = 2.4
Identities = 29/98 (29%), Positives = 34/98 (34%), Gaps = 5/98 (5%)
Frame = +1
Query: 20 EGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGG 79
+GGG +GGGG R VA + G + GGG GG GG
Sbjct: 529 DGGGGTGGGG-------RGGRGYVATRTRGTSAGRGCPPIFLARGTRSHSGGGGGGRGGG 687
Query: 80 --GARAWWGRRRQWWRWRQWCDDG---GGSGSDDGGGS 112
G A R Q G GG+G DG S
Sbjct: 688 RRGVAAGCARGMLCGARAQGAPGGVALGGAGRADGARS 801
Score = 25.4 bits (54), Expect = 5.3
Identities = 32/121 (26%), Positives = 38/121 (30%), Gaps = 26/121 (21%)
Frame = +2
Query: 21 GGGDSGGGGW---C---------WWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDG---- 64
GGG GGG W C W R R V V E + + G
Sbjct: 176 GGGPGGGGRWRGGCPRPQAAC*GEWGRGRGGGPVQGRRGAGWVGEGGAGDGALDPGVPPL 355
Query: 65 ----------GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG 114
DGG +GG G G +W GG +G GGG+GG
Sbjct: 356 GSFLTLARAPRRRPDGGRAGGRRGRG---------------RWGAGGGRAGGRRGGGAGG 490
Query: 115 G 115
G
Sbjct: 491 G 493
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 42.0 bits (97), Expect(2) = 2e-06
Identities = 24/58 (41%), Positives = 26/58 (44%)
Frame = +1
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGG N GGG GGG G + GGG G GGG GGGG GG
Sbjct: 319 NGGGYNHGGGGYNHGGGGYNHGGGGY----------NGGGGHGGHGGGGYNGGGGHGG 462
Score = 39.7 bits (91), Expect = 3e-04
Identities = 23/61 (37%), Positives = 30/61 (48%), Gaps = 3/61 (4%)
Frame = +1
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQ---WCDDGGGSGSDDGGGSGGGGGGG 119
+GGGG + GG G ++GGG G + + GGG + GGG GGGG G
Sbjct: 238 NGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHG 417
Query: 120 G 120
G
Sbjct: 418 G 420
Score = 38.5 bits (88), Expect = 6e-04
Identities = 24/61 (39%), Positives = 29/61 (47%), Gaps = 3/61 (4%)
Frame = +1
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG---GGG 119
+GGG N GGG + GGG G ++GGG + GGG GGG GGG
Sbjct: 199 NGGGYNHGGGGYNNGGGGYNHGGG----------GYNNGGGGYNHGGGGYNGGGYNHGGG 348
Query: 120 G 120
G
Sbjct: 349 G 351
Score = 38.5 bits (88), Expect = 6e-04
Identities = 33/100 (33%), Positives = 36/100 (36%)
Frame = +1
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG +GGGG A +V V E GGG N GGGS GG
Sbjct: 427 GGGYNGGGG--------HGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGS 582
Query: 81 ARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG G + GGG GG GGGG
Sbjct: 583 YHH------------------GGGGYNHGGGGHGGHGGGG 648
Score = 36.6 bits (83), Expect = 0.002
Identities = 26/69 (37%), Positives = 33/69 (47%), Gaps = 11/69 (15%)
Frame = +1
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRR-----RQWWRWRQWCDDGGGSGSDDGGGS---GG 114
+GGGG+ G G GG +GGG G + + + D G G + GGGS GG
Sbjct: 397 NGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGG 576
Query: 115 GG---GGGG 120
G GGGG
Sbjct: 577 GSYHHGGGG 603
Score = 34.3 bits (77), Expect = 0.011
Identities = 22/55 (40%), Positives = 26/55 (47%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
GGG N GG GG +GGG G ++GGG + GGG GGGG
Sbjct: 166 GGGYNHGG--GGYNGGGYNHGGG----------GYNNGGGGYNHGGGGYNNGGGG 294
Score = 28.1 bits (61), Expect = 0.82
Identities = 12/23 (52%), Positives = 12/23 (52%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWG 86
GGG N GGG G GGG G
Sbjct: 595 GGGYNHGGGGHGGHGGGGHGGHG 663
Score = 26.2 bits (56), Expect = 3.1
Identities = 7/14 (50%), Positives = 8/14 (57%)
Frame = +3
Query: 84 WWGRRRQWWRWRQW 97
W R WWRW +W
Sbjct: 615 WRWTRGTWWRWSRW 656
Score = 24.3 bits (51), Expect(2) = 2e-06
Identities = 11/24 (45%), Positives = 17/24 (70%)
Frame = +2
Query: 39 SVVVVAVTVVVIVVEAVVVEAVVV 62
++V V TVVV+++ V+ AVVV
Sbjct: 161 AMVEVTTTVVVVIMVEVITTAVVV 232
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 46.6 bits (109), Expect = 2e-06
Identities = 29/67 (43%), Positives = 29/67 (43%), Gaps = 9/67 (13%)
Frame = -1
Query: 63 DGGGGNDGGG------SGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG 116
DGGG DGGG G GGG G GGG GS GGG GGGG
Sbjct: 405 DGGGDGDGGG**GV*CGAGDDGGGGSTQGG-------------GGGGGGSTQGGGDGGGG 265
Query: 117 ---GGGG 120
GGGG
Sbjct: 264 E*TGGGG 244
Score = 40.0 bits (92), Expect = 2e-04
Identities = 25/66 (37%), Positives = 26/66 (38%), Gaps = 9/66 (13%)
Frame = -1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGS---------DDGGGSGG 114
GGGG G GG GGG G C GGG G + GGG G
Sbjct: 318 GGGGGGGSTQGGGDGGGGE*TGGGGE--------CTGGGGGGDKYGYTGGVGE*GGGGGE 163
Query: 115 GGGGGG 120
GGGGG
Sbjct: 162 XGGGGG 145
Score = 38.5 bits (88), Expect = 6e-04
Identities = 26/61 (42%), Positives = 28/61 (45%), Gaps = 4/61 (6%)
Frame = -1
Query: 64 GGGGNDGGGSGGSSGGGARAWW--GRRRQWWRWRQWCDDGGGSGSDDGGGSGGG--GGGG 119
GGGG GGG G +GGG G GGG + GGG GGG GGGG
Sbjct: 177 GGGGEXGGGGGE*AGGGGGEL***GT------------GGGGENTGAGGGGGGGEFGGGG 34
Query: 120 G 120
G
Sbjct: 33 G 31
Score = 31.6 bits (70), Expect = 0.074
Identities = 26/73 (35%), Positives = 30/73 (40%), Gaps = 16/73 (21%)
Frame = -1
Query: 64 GGGGNDG------GGSGGS--SGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG-- 113
GGGG D GG G S GGG + G + + + DGGG GGG G
Sbjct: 585 GGGGGDAYEIPKTGGIGPSYTGGGGGDEYTGGGGEGYAYT---GDGGGEVYTGGGGEG*A 415
Query: 114 ------GGGGGGG 120
G G GGG
Sbjct: 414 YTGDGGGDGDGGG 376
Score = 28.1 bits (61), Expect = 0.82
Identities = 10/20 (50%), Positives = 11/20 (55%), Gaps = 4/20 (20%)
Frame = -3
Query: 82 RAWWGRRRQWWR----WRQW 97
R WW R R WWR W +W
Sbjct: 181 RWWWWRXRWWWR*VSWWWRW 122
Score = 26.9 bits (58), Expect(2) = 0.29
Identities = 11/23 (47%), Positives = 12/23 (51%)
Frame = -1
Query: 64 GGGGNDGGGSGGSSGGGARAWWG 86
G GG GGG G GGG +G
Sbjct: 78 GAGGGGGGGEFGGGGGGDLTQYG 10
Score = 21.2 bits (43), Expect(2) = 0.29
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = -2
Query: 39 SVVVVAVTVVVIVVEAVVVEAV 60
S VVV VV+ V +VVE V
Sbjct: 185 SEVVVVANXVVVEVSELVVEVV 120
>BQ140560 weakly similar to GP|15128223|db hypothetical protein~similar to
Oryza sativa chromosome 1 P0489A01.2, partial (5%)
Length = 215
Score = 45.4 bits (106), Expect = 5e-06
Identities = 25/53 (47%), Positives = 27/53 (50%)
Frame = -1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG 116
GGGG +GGG GG G GA GR D GGG G+ GG GGGG
Sbjct: 143 GGGGIEGGGGGGGGGRGAPLDAGR----------YDGGGGGGAP*NGGGGGGG 15
Score = 40.4 bits (93), Expect = 2e-04
Identities = 22/56 (39%), Positives = 24/56 (42%)
Frame = -1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG S GGG W +GGG G +GGG GGGGG G
Sbjct: 215 GGGGAPVDAGRYSXGGGGAPW---------------NGGGGGGIEGGGGGGGGGRG 93
Score = 32.0 bits (71), Expect = 0.057
Identities = 20/63 (31%), Positives = 24/63 (37%)
Frame = -1
Query: 19 VEGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSG 78
+EGGG GGGG ++A DGGGG +GG G
Sbjct: 131 IEGGGGGGGGG-----------------------RGAPLDAGRYDGGGGGGAP*NGGGGG 21
Query: 79 GGA 81
GGA
Sbjct: 20 GGA 12
Score = 29.6 bits (65), Expect = 0.28
Identities = 18/43 (41%), Positives = 21/43 (47%)
Frame = -1
Query: 78 GGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGA GR + GGG+ + GGG G GGGGG
Sbjct: 212 GGGAPVDAGR---------YSXGGGGAPWNGGGGGGIEGGGGG 111
>TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like
{Arabidopsis thaliana}, partial (87%)
Length = 1177
Score = 44.3 bits (103), Expect = 1e-05
Identities = 25/57 (43%), Positives = 28/57 (48%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG G GG GG GGG R + G R ++ R GG GGG G GGG GG
Sbjct: 69 GGFGGRGGDRGGRGGGGGRGFGGGRGGDFKPRGGGRGFGGGRGARGGGRGRGGGRGG 239
Score = 25.4 bits (54), Expect = 5.3
Identities = 10/16 (62%), Positives = 10/16 (62%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGG 79
GGG GGG GG GG
Sbjct: 204 GGGRGRGGGRGGMKGG 251
>AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (10%)
Length = 478
Score = 43.1 bits (100), Expect = 2e-05
Identities = 29/62 (46%), Positives = 32/62 (50%), Gaps = 4/62 (6%)
Frame = -3
Query: 63 DGGGGNDGGGSGGSSGG-GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG---GGGGG 118
+G GG+ GGG G + GG GA G W C GGG DGGG G GGGGG
Sbjct: 374 EGEGGSGGGGDGRTCGGEGAIG*CGDGGGWL-----CG-GGGENG*DGGGGGXLRGGGGG 213
Query: 119 GG 120
GG
Sbjct: 212 GG 207
Score = 38.5 bits (88), Expect = 6e-04
Identities = 25/64 (39%), Positives = 29/64 (45%), Gaps = 7/64 (10%)
Frame = -3
Query: 64 GGGGNDGGGSGGSS-----GGGARAWWGRRRQWWRWRQWCDDGGG--SGSDDGGGSGGGG 116
GGGG++ G+GG GGG + W G DGGG DGGG G
Sbjct: 218 GGGGDE*DGAGGGDLME*GGGGLKGWKG-------------DGGGLMGWKGDGGGLRGWE 78
Query: 117 GGGG 120
GGGG
Sbjct: 77 GGGG 66
Score = 34.7 bits (78), Expect = 0.009
Identities = 25/63 (39%), Positives = 27/63 (42%), Gaps = 6/63 (9%)
Frame = -3
Query: 63 DGGGGN-DGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGG-----G 116
DGGG GGG G GGG G GGG G D+ G+GGG G
Sbjct: 299 DGGGWLCGGGGENG*DGGGGGXLRG--------------GGGGGGDE*DGAGGGDLME*G 162
Query: 117 GGG 119
GGG
Sbjct: 161 GGG 153
Score = 25.8 bits (55), Expect = 4.1
Identities = 9/16 (56%), Positives = 10/16 (62%)
Frame = -2
Query: 84 WWGRRRQWWRWRQWCD 99
W GRR +WW W W D
Sbjct: 378 WRGRRWKWW-WWGWAD 334
Score = 25.8 bits (55), Expect(2) = 0.41
Identities = 14/29 (48%), Positives = 15/29 (51%), Gaps = 4/29 (13%)
Frame = -3
Query: 63 DGGG----GNDGGGSGGSSGGGARAWWGR 87
DGGG DGGG G GGG R G+
Sbjct: 134 DGGGLMGWKGDGGGLRGWEGGGGRCGNGK 48
Score = 21.9 bits (45), Expect(2) = 0.41
Identities = 5/7 (71%), Positives = 6/7 (85%)
Frame = -2
Query: 30 WCWWRRW 36
WCWW R+
Sbjct: 195 WCWWWRF 175
>TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (58%)
Length = 612
Score = 40.4 bits (93), Expect = 2e-04
Identities = 27/67 (40%), Positives = 29/67 (42%), Gaps = 10/67 (14%)
Frame = +2
Query: 64 GGGGNDGGGSGG----SSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG--- 116
GGGG GGG GG GGG ++ GGG G GGG GGGG
Sbjct: 308 GGGGRGGGGYGGGYDQGYGGGGGSY----------------GGGFGGGRGGGRGGGGYXQ 439
Query: 117 ---GGGG 120
GGGG
Sbjct: 440 GGYGGGG 460
Score = 40.4 bits (93), Expect = 2e-04
Identities = 27/58 (46%), Positives = 29/58 (49%), Gaps = 1/58 (1%)
Frame = +2
Query: 64 GGGGNDG-GGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG + G GG GGS GGG G R + Q GGG G GG G GG GGG
Sbjct: 338 GGGYDQGYGGGGGSYGGGFGGGRGGGRGGGGYXQG-GYGGGGGYXQGGYGGQGGYGGG 508
Score = 38.9 bits (89), Expect = 5e-04
Identities = 24/57 (42%), Positives = 26/57 (45%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG GG + + G GG G GGG GGG GGGG
Sbjct: 305 GGGGGRGG--GGYGGGYDQGYGG-------------GGGSYGGGFGGGRGGGRGGGG 430
Score = 30.8 bits (68), Expect = 0.13
Identities = 19/51 (37%), Positives = 19/51 (37%)
Frame = +2
Query: 70 GGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G GG GGG G G G D G G GGG GGG
Sbjct: 299 GYGGGGGRGGG--------------------GYGGGYDQGYGGGGGSYGGG 391
>TC93504 weakly similar to PIR|S46286|S46286 RNA-binding protein - wood
tobacco, partial (55%)
Length = 576
Score = 38.5 bits (88), Expect = 6e-04
Identities = 22/49 (44%), Positives = 24/49 (48%), Gaps = 7/49 (14%)
Frame = +3
Query: 64 GGGGNDG-----GGSGGSSGGGAR--AWWGRRRQWWRWRQWCDDGGGSG 105
GGGG G GG GG SGGG WW R+ WWR + GG G
Sbjct: 411 GGGGGYGGGGGYGGGGGYSGGGGNYGGWW--RKLWWRKLWVAETTGGVG 551
Score = 37.7 bits (86), Expect = 0.001
Identities = 20/54 (37%), Positives = 29/54 (53%)
Frame = +1
Query: 39 SVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWW 92
+ V +A V ++VV A+VV ++ GGGGN GGG+ G W + R W
Sbjct: 415 AAVAMAAAVAMVVVVAIVVVVEIMVGGGGNYGGGNYG---------WRKLRGGW 549
Score = 35.4 bits (80), Expect = 0.005
Identities = 19/40 (47%), Positives = 26/40 (64%)
Frame = +1
Query: 3 VAMMVAAAVVVAMLAVVEGGGDSGGGGWCWWRRWRRSVVV 42
VAM+V A+VV + +V GGG+ GGG + WR+ R VV
Sbjct: 439 VAMVVVVAIVVVVEIMVGGGGNYGGGNY-GWRKLRGGWVV 555
Score = 32.0 bits (71), Expect = 0.057
Identities = 15/22 (68%), Positives = 15/22 (68%), Gaps = 2/22 (9%)
Frame = +3
Query: 101 GGGSGSDDGGGSGGGGG--GGG 120
GGG G GGG GGGGG GGG
Sbjct: 411 GGGGGYGGGGGYGGGGGYSGGG 476
Score = 31.6 bits (70), Expect = 0.074
Identities = 22/56 (39%), Positives = 22/56 (39%)
Frame = +3
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG G GGG GGG GG SGGGG GG
Sbjct: 396 GGGGYGGGGGYGGGGGY-------------------GGG-----GGYSGGGGNYGG 491
Score = 31.2 bits (69), Expect = 0.097
Identities = 13/19 (68%), Positives = 13/19 (68%)
Frame = +3
Query: 102 GGSGSDDGGGSGGGGGGGG 120
GG G GGG GGGGG GG
Sbjct: 396 GGGGYGGGGGYGGGGGYGG 452
Score = 28.1 bits (61), Expect = 0.82
Identities = 12/35 (34%), Positives = 15/35 (42%)
Frame = +2
Query: 84 WWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
WW WW W+ W ++ GG GGGG
Sbjct: 446 WWWWWL*WWWWKLWWVVAEIMVAEIMGGGNYGGGG 550
Score = 28.1 bits (61), Expect = 0.82
Identities = 15/29 (51%), Positives = 17/29 (57%), Gaps = 3/29 (10%)
Frame = +3
Query: 21 GGGDSGGGG--WCWWRR-WRRSVVVVAVT 46
GGG SGGGG WWR+ W R + V T
Sbjct: 453 GGGYSGGGGNYGGWWRKLWWRKLWVAETT 539
Score = 26.2 bits (56), Expect = 3.1
Identities = 19/52 (36%), Positives = 24/52 (45%), Gaps = 9/52 (17%)
Frame = +2
Query: 30 WCWWRR--WRRSVV-------VVAVTVVVIVVEAVVVEAVVVDGGGGNDGGG 72
W WWRR WRR + + +V E +V E + GGGN GGG
Sbjct: 404 WLWWRRWLWRRRWLWWWWWL*WWWWKLWWVVAEIMVAEIM----GGGNYGGG 547
Score = 24.6 bits (52), Expect = 9.0
Identities = 16/45 (35%), Positives = 17/45 (37%), Gaps = 9/45 (20%)
Frame = +2
Query: 82 RAW-WGRRRQWWRW--------RQWCDDGGGSGSDDGGGSGGGGG 117
R W W RR WW W W GGG+ GGGG
Sbjct: 416 RRWLWRRRWLWWWWWL*WWWWKLWWVVAEIMVAEIMGGGNYGGGG 550
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.142 0.499
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,275,596
Number of Sequences: 36976
Number of extensions: 94735
Number of successful extensions: 12525
Number of sequences better than 10.0: 786
Number of HSP's better than 10.0 without gapping: 3312
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7472
length of query: 120
length of database: 9,014,727
effective HSP length: 84
effective length of query: 36
effective length of database: 5,908,743
effective search space: 212714748
effective search space used: 212714748
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0090b.2


