

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
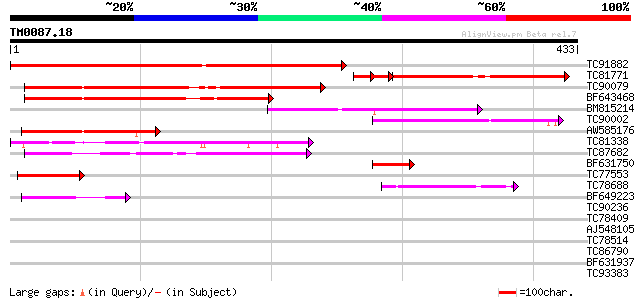
Query= TM0087.18
(433 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91882 similar to GP|10176714|dbj|BAB09944. dimethylaniline mon... 390 e-109
TC81771 similar to GP|10176714|dbj|BAB09944. dimethylaniline mon... 188 2e-56
TC90079 similar to GP|5454204|gb|AAD43619.1| T3P18.18 {Arabidops... 193 1e-49
BF643468 weakly similar to PIR|F96682|F96 hypothetical protein F... 167 6e-42
BM815214 similar to GP|22136992|gb unknown protein {Arabidopsis ... 135 3e-32
TC90002 weakly similar to GP|9757850|dbj|BAB08484.1 dimethylanil... 128 5e-30
AW585176 similar to GP|14532474|gb| At1g62600/T3P18_16 {Arabidop... 117 1e-26
TC81338 weakly similar to PIR|B86326|B86326 hypothetical protein... 79 3e-15
TC87682 weakly similar to PIR|T04527|T04527 hypothetical protein... 78 6e-15
BF631750 similar to GP|10176714|dbj dimethylaniline monooxygenas... 55 4e-08
TC77553 homologue to GP|1066499|gb|AAB41904.1| NADH-dependent gl... 47 1e-05
TC78688 weakly similar to PIR|B86326|B86326 hypothetical protein... 42 5e-04
BF649223 weakly similar to GP|21740857|emb OSJNBb0072N21.6 {Oryz... 42 6e-04
TC90236 similar to GP|4586308|dbj|BAA76348.1 protoporphyrinogen ... 37 0.019
TC78409 homologue to GP|18086418|gb|AAL57665.1 AT3g59050/F17J16_... 36 0.033
AJ548105 35 0.074
TC78514 homologue to SP|Q9SMJ3|ZDS_CAPAN Zeta-carotene desaturas... 33 0.21
TC86790 similar to PIR|A84861|A84861 probable amine oxidase [imp... 33 0.28
BF631937 similar to GP|10177621|db phytoene dehydrogenase-like {... 33 0.28
TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16... 33 0.28
>TC91882 similar to GP|10176714|dbj|BAB09944. dimethylaniline
monooxygenase-like protein {Arabidopsis thaliana},
partial (47%)
Length = 800
Score = 390 bits (1002), Expect = e-109
Identities = 194/260 (74%), Positives = 220/260 (84%), Gaps = 3/260 (1%)
Frame = +2
Query: 1 MVSKTNQ-NSKNVCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQWLYNP-NVQCED 58
MVS+TNQ SKNVCVIGAGP+GLVA RE+++EGHKV+V EQNHDIGGQWLY+ N++ ED
Sbjct: 26 MVSETNQYQSKNVCVIGAGPAGLVAAREIRKEGHKVIVLEQNHDIGGQWLYDDKNIEGED 205
Query: 59 PLGKNPVLKVHSSIYESLRLMSPREIMGFTDFPFLVKKGRDMRRFPGHRELLFYLNDFCE 118
PLG+NP LKVHSSIY SLR SPRE+MGFTDFPF KKGRDMRRFPGH E+L YL DFCE
Sbjct: 206 PLGRNPFLKVHSSIYNSLRTQSPREVMGFTDFPFSTKKGRDMRRFPGHVEILMYLKDFCE 385
Query: 119 WFGLREVIRFNTRVEYVGPLNYNGAFSGNDDLKWVVRSKEKESDGVVEEVFDAVVVASGH 178
WFGLRE+IRFNTRV YVG L+ G N LKW+VRSKEK SD VVEEVFDAVVVA+GH
Sbjct: 386 WFGLREMIRFNTRVNYVGMLDDYGVCGNN--LKWIVRSKEKNSDKVVEEVFDAVVVATGH 559
Query: 179 YSHPKLPCIQGMDTWKRKQMHSHIYRTPQPFGKEIVVVVGNSFSGQEISMDLVKVAKEVH 238
Y PKLP I+GMDTWKRKQMHSHIYRTP+PF E+VVVVGNSFSGQ+I+++LV VAKEVH
Sbjct: 560 YCQPKLPSIKGMDTWKRKQMHSHIYRTPEPFHNEVVVVVGNSFSGQDIAIELVGVAKEVH 739
Query: 239 LSSNSA-YISEGLSKVIHKY 257
+SS S I+EG SKVI K+
Sbjct: 740 ISSRSLDTITEGXSKVISKH 799
>TC81771 similar to GP|10176714|dbj|BAB09944. dimethylaniline
monooxygenase-like protein {Arabidopsis thaliana},
partial (39%)
Length = 874
Score = 188 bits (477), Expect(3) = 2e-56
Identities = 90/135 (66%), Positives = 109/135 (80%)
Frame = +2
Query: 293 PSLAPSLSFMGIPSKVLGPHFFESQAKWIAQLLSGKKILPSFEEMMKSIKELYHSREVAN 352
P L+PSL+F+GIP K+LG FFESQA WIAQLLSGKK+LPS+EEMMKSI+E YH REVA
Sbjct: 188 PLLSPSLTFIGIPKKLLGFPFFESQAMWIAQLLSGKKVLPSWEEMMKSIEEFYHEREVAG 367
Query: 353 VPKRYTHSIADNDGFEYFDKYGDNAGFPKLEEWRKEICISSTVNMFANLETFRDSWNDDE 412
+P TH IA+ FEY DKY +N G P L+EWRK +C+S+ +N F NLET+RDSW+DDE
Sbjct: 368 IP---THDIAN---FEYCDKYSENVGLPPLQEWRKVLCVSAIINSFVNLETYRDSWDDDE 529
Query: 413 MLQEVLKIPYFTQLG 427
+LQE L+ PYFTQLG
Sbjct: 530 VLQEALQSPYFTQLG 574
Score = 42.0 bits (97), Expect(3) = 2e-56
Identities = 16/18 (88%), Positives = 17/18 (93%)
Frame = +1
Query: 276 WFIEDNRVGPLYEHTFPP 293
W IED+RVGPLYEHTFPP
Sbjct: 136 WVIEDDRVGPLYEHTFPP 189
Score = 28.5 bits (62), Expect(3) = 2e-56
Identities = 11/17 (64%), Positives = 14/17 (81%)
Frame = +3
Query: 263 HPQIDNLQEDGREWFIE 279
HPQID L+EDGR F++
Sbjct: 3 HPQIDTLEEDGRVIFMD 53
>TC90079 similar to GP|5454204|gb|AAD43619.1| T3P18.18 {Arabidopsis
thaliana}, partial (38%)
Length = 677
Score = 193 bits (490), Expect = 1e-49
Identities = 104/231 (45%), Positives = 144/231 (62%), Gaps = 1/231 (0%)
Frame = +3
Query: 12 VCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQWLYNPNVQCEDPLGKNPVLK-VHS 70
V VIGAG SG+VA +EL++EGH V+VFE+N+ +GG W+Y DPL +P + VHS
Sbjct: 21 VAVIGAGVSGMVAAKELQQEGHNVIVFEKNNRVGGTWIYTSKSD-SDPLSIDPTRETVHS 197
Query: 71 SIYESLRLMSPREIMGFTDFPFLVKKGRDMRRFPGHRELLFYLNDFCEWFGLREVIRFNT 130
S+Y SLR PR IMGF D+P ++ D R FPGH E+L +L +F + FG+ E+ RF T
Sbjct: 198 SVYLSLRTNLPRHIMGFLDYPLSKRESGDPRTFPGHEEVLRFLEEFADEFGIHELTRFET 377
Query: 131 RVEYVGPLNYNGAFSGNDDLKWVVRSKEKESDGVVEEVFDAVVVASGHYSHPKLPCIQGM 190
V V G D WVV S+ D V EVF+AVVV SGHY P+L + G+
Sbjct: 378 EVVKVER-------KGGKD--WVVESRG--GDSVSREVFEAVVVCSGHYVEPRLAVVPGI 524
Query: 191 DTWKRKQMHSHIYRTPQPFGKEIVVVVGNSFSGQEISMDLVKVAKEVHLSS 241
+ + QMHSH YR P F ++V+++G S +IS D+ ++AKEVH+++
Sbjct: 525 ENFGGFQMHSHNYRVPHSFKDQVVILIGLGTSSFDISRDIARLAKEVHVAT 677
>BF643468 weakly similar to PIR|F96682|F96 hypothetical protein F12P19.2
[imported] - Arabidopsis thaliana, partial (33%)
Length = 655
Score = 167 bits (424), Expect = 6e-42
Identities = 89/191 (46%), Positives = 120/191 (62%), Gaps = 1/191 (0%)
Frame = +2
Query: 12 VCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQWLYNPNVQCEDPLGKNPVLK-VHS 70
V VIGAG SGLVA +EL++EGH VVVFE+N+ +GG W+Y P DPL +P + VHS
Sbjct: 89 VAVIGAGVSGLVAAKELQQEGHNVVVFEKNNRVGGTWIYTPKTD-SDPLSMDPARETVHS 265
Query: 71 SIYESLRLMSPREIMGFTDFPFLVKKGRDMRRFPGHRELLFYLNDFCEWFGLREVIRFNT 130
S+Y SLR PR+IMGF D+P ++ D R FPGH E+L +L +F FG+ E+ RF T
Sbjct: 266 SVYHSLRTNLPRQIMGFLDYPLSKRESGDPRTFPGHEEVLRFLEEFAGEFGIHELTRFET 445
Query: 131 RVEYVGPLNYNGAFSGNDDLKWVVRSKEKESDGVVEEVFDAVVVASGHYSHPKLPCIQGM 190
V V + +W+V S + D V +EVF+AVVV SGH+ P+L + G+
Sbjct: 446 EVVKV----------ERKENEWIVES--RGGDSVSQEVFEAVVVCSGHFVEPRLAVVPGI 589
Query: 191 DTWKRKQMHSH 201
+T+ QMHSH
Sbjct: 590 ETFPGFQMHSH 622
>BM815214 similar to GP|22136992|gb unknown protein {Arabidopsis thaliana},
partial (24%)
Length = 596
Score = 135 bits (340), Expect = 3e-32
Identities = 78/195 (40%), Positives = 108/195 (55%), Gaps = 31/195 (15%)
Frame = +1
Query: 198 MHSHIYRTPQPFGKEIVVVVGNSFSGQEISMDLVKVAKEVHLSSNSAYISEGLSKVIHKY 257
MHSH YR P F ++V+++G S +IS D+ VAKEVH+++ + +G+ +
Sbjct: 13 MHSHNYRVPHSFKDQVVILIGLGPSSFDISKDIAGVAKEVHVATKPNPLLKGMK--LENV 186
Query: 258 QNFHLHPQIDNLQEDGREWF-------------------------------IEDNRVGPL 286
+N H I + EDG F IEDNRVGPL
Sbjct: 187 RNICFHTLIKCVYEDGLVAFEDGFSTYADAIIHCTGYKYHIPFLETNGIVTIEDNRVGPL 366
Query: 287 YEHTFPPSLAPSLSFMGIPSKVLGPHFFESQAKWIAQLLSGKKILPSFEEMMKSIKELYH 346
Y+H FPPSLAP LSF+G+ + E QAKW+A++LSGK +LP+ EEMM+S+K++Y
Sbjct: 367 YKHVFPPSLAPGLSFIGLTFRETIFVVIELQAKWVARVLSGKILLPTEEEMMESVKDMYR 546
Query: 347 SREVANVPKRYTHSI 361
E +PKRYTHSI
Sbjct: 547 VMEENALPKRYTHSI 591
>TC90002 weakly similar to GP|9757850|dbj|BAB08484.1 dimethylaniline
monooxygenase (N-oxide-forming)-like protein
{Arabidopsis thaliana}, partial (26%)
Length = 778
Score = 128 bits (321), Expect = 5e-30
Identities = 71/157 (45%), Positives = 92/157 (58%), Gaps = 11/157 (7%)
Frame = +3
Query: 278 IEDNRVGPLYEHTFPPSLAPSLSFMGIPSKVLGPHFFESQAKWIAQLLSGKKILPSFEEM 337
IEDNRVGPLY+H FPPSLAP LSF+G+ K + E QAKWIA++LSGK +LP EEM
Sbjct: 189 IEDNRVGPLYKHVFPPSLAPCLSFIGLTFKEITFSVIELQAKWIARVLSGKVLLPDEEEM 368
Query: 338 MKSIKELYHSREVANVPKRYTHSIADNDGFEYFDKYGDNAGFPKLEEWRKEICISSTVNM 397
M SIK+ Y S E + KR THS+ G +Y + G P LE+WR + + N
Sbjct: 369 MASIKDFYQSMEENGLSKRQTHSLRPLQG-DYKHWLVEQIGLPPLEDWRDNMLMECLKNS 545
Query: 398 FANLETFRDSWND---DEMLQ--------EVLKIPYF 423
E FRD W+D D ++Q + L +P+F
Sbjct: 546 IEMNEMFRDEWDDNYWDAIIQSGSAS*QAKTLNVPFF 656
>AW585176 similar to GP|14532474|gb| At1g62600/T3P18_16 {Arabidopsis
thaliana}, partial (21%)
Length = 476
Score = 117 bits (292), Expect = 1e-26
Identities = 61/109 (55%), Positives = 75/109 (67%), Gaps = 3/109 (2%)
Frame = +3
Query: 10 KNVCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQWLYNPNVQCEDPLGKNPVLK-V 68
++V IGAG GLVA REL+REGH+VVVFE+ ++GG W+Y V+ DPLG +P K +
Sbjct: 153 RHVFFIGAGAGGLVAARELRREGHQVVVFERGEELGGSWVYTSEVE-SDPLGLDPNRKLI 329
Query: 69 HSSIYESLRLMSPREIMGFTDFPFLVK--KGRDMRRFPGHRELLFYLND 115
HSS+Y SLR PRE MGF D+PF K KGRD RFP H E+L Y D
Sbjct: 330 HSSLYNSLRTNLPRESMGFRDYPFRRKEEKGRDSTRFPSHGEVLMYSKD 476
>TC81338 weakly similar to PIR|B86326|B86326 hypothetical protein AAF82235.1
[imported] - Arabidopsis thaliana, partial (32%)
Length = 784
Score = 79.0 bits (193), Expect = 3e-15
Identities = 70/255 (27%), Positives = 113/255 (43%), Gaps = 23/255 (9%)
Frame = +2
Query: 1 MVSKTNQNS--KNVCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQWLYNPNVQCED 58
M +K N S + +IGAG SGL ++L + +VFE IGG W + C
Sbjct: 32 MPTKDNPPSIVSRIGIIGAGVSGLAVAKQLSH--YNPIVFEATDSIGGVWRH-----CS- 187
Query: 59 PLGKNPVLKVHSSIYESLRLMSPREIMGFTDFPFLVKKGRDMRRFPGHRELLFYLNDFCE 118
Y +L S F+DFP+ ++ +D +P H E+L YL+ +
Sbjct: 188 --------------YRCTKLQSQTWNYEFSDFPWPERESKD---YPSHAEILEYLHLYAV 316
Query: 119 WFGLREVIRFNTRVEYVGPLNYNGAFS--------GN---DDLKWVVRSKEKESDGVVEE 167
+F L + ++FNT+V + + F GN + W + + ESD + +
Sbjct: 317 YFDLFKYVKFNTKVVEIKFVRDKEGFDFGRLPGDHGNPLPERPVWELSVQTNESDAIQKY 496
Query: 168 VFDAVVVASGHYSH----PKLPCIQGMDTWKRKQMHSHIY------RTPQPFGKEIVVVV 217
F+ VVV +G Y P PC +G + +K K +H+ Y T + VVVV
Sbjct: 497 CFEFVVVCTGKYGDIPLMPTFPCNKGPEVFKGKVLHTIDYCKLDKEATSDLVKGKKVVVV 676
Query: 218 GNSFSGQEISMDLVK 232
G S +++M+ V+
Sbjct: 677 GYKKSAIDLTMECVQ 721
>TC87682 weakly similar to PIR|T04527|T04527 hypothetical protein F16A16.170
- Arabidopsis thaliana, partial (38%)
Length = 1040
Score = 78.2 bits (191), Expect = 6e-15
Identities = 55/219 (25%), Positives = 99/219 (45%)
Frame = +1
Query: 12 VCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQWLYNPNVQCEDPLGKNPVLKVHSS 71
V +IGAG SGL L ++ ++ E+ + W +
Sbjct: 43 VIIIGAGTSGLATAACLTKQSIPFIILERENCFASLW--------------------QNY 162
Query: 72 IYESLRLMSPREIMGFTDFPFLVKKGRDMRRFPGHRELLFYLNDFCEWFGLREVIRFNTR 131
Y+ + L +++ FPF + ++ + YL ++ F + + +N
Sbjct: 163 TYDRVHLHLRKQLCELPHFPF----PPSYPHYVPKKQFIEYLGNYVNNFNINPI--YNRA 324
Query: 132 VEYVGPLNYNGAFSGNDDLKWVVRSKEKESDGVVEEVFDAVVVASGHYSHPKLPCIQGMD 191
VE + +D+ KW V+++ K S V E +VVASG + P++P ++G++
Sbjct: 325 VELA-------EYVDDDEKKWRVKAENKSSGEVEEYSARFLVVASGETAEPRVPVVEGLE 483
Query: 192 TWKRKQMHSHIYRTPQPFGKEIVVVVGNSFSGQEISMDL 230
+K K +HS Y+ + F E V+VVG+ SG EI++DL
Sbjct: 484 NFKGKVIHSTRYKNGKEFKDEHVLVVGSGNSGMEIALDL 600
>BF631750 similar to GP|10176714|dbj dimethylaniline monooxygenase-like
protein {Arabidopsis thaliana}, partial (11%)
Length = 558
Score = 55.5 bits (132), Expect = 4e-08
Identities = 24/32 (75%), Positives = 29/32 (90%)
Frame = +2
Query: 278 IEDNRVGPLYEHTFPPSLAPSLSFMGIPSKVL 309
IED+RVGPLYEHTF P L+PSL+F+GIP KV+
Sbjct: 59 IEDDRVGPLYEHTFHPLLSPSLTFIGIPKKVI 154
>TC77553 homologue to GP|1066499|gb|AAB41904.1| NADH-dependent glutamate
synthase {Medicago sativa}, partial (37%)
Length = 3067
Score = 47.0 bits (110), Expect = 1e-05
Identities = 22/52 (42%), Positives = 33/52 (63%), Gaps = 1/52 (1%)
Frame = +1
Query: 7 QNSKNVCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQWLYN-PNVQCE 57
+ K V ++G+GPSGL A +L + GH V VFE+ IGG +Y PN++ +
Sbjct: 1366 RTGKRVAIVGSGPSGLAAADQLNKMGHIVTVFERADRIGGLMMYGVPNMKTD 1521
>TC78688 weakly similar to PIR|B86326|B86326 hypothetical protein AAF82235.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1294
Score = 42.0 bits (97), Expect = 5e-04
Identities = 30/105 (28%), Positives = 55/105 (51%), Gaps = 1/105 (0%)
Frame = +3
Query: 285 PLYEHTFPPSLAPSLSFMGIPSKVLGPHFFESQAKWIAQLLSGKKILPSFEEMM-KSIKE 343
PLY T PS+ P+++F+G V + E ++ W+A L+ K LPS E+M+ ++IK+
Sbjct: 708 PLYRGTVHPSI-PNMAFVGYVESVSNLYTSEIRSMWLAGLIDNKFKLPSAEKMLSQTIKD 884
Query: 344 LYHSREVANVPKRYTHSIADNDGFEYFDKYGDNAGFPKLEEWRKE 388
+ ++ KR + G + D+ ++ G+ WRK+
Sbjct: 885 METMKKSTRFYKR---NCITTFGINHNDEICEDLGW---HTWRKK 1001
>BF649223 weakly similar to GP|21740857|emb OSJNBb0072N21.6 {Oryza sativa},
partial (11%)
Length = 278
Score = 41.6 bits (96), Expect = 6e-04
Identities = 23/83 (27%), Positives = 39/83 (46%)
Frame = +2
Query: 10 KNVCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQWLYNPNVQCEDPLGKNPVLKVH 69
K V ++GAG SGL+A + + + G +VFE + +GG W +
Sbjct: 71 KRVAIVGAGISGLLACKYVTQIGFNPIVFEADDGVGGLWRHT------------------ 196
Query: 70 SSIYESLRLMSPREIMGFTDFPF 92
ES +L +P++ F+ FP+
Sbjct: 197 ---IESTKLQNPKQAYQFSXFPW 256
>TC90236 similar to GP|4586308|dbj|BAA76348.1 protoporphyrinogen IX
oxidase {Glycine max}, partial (39%)
Length = 686
Score = 36.6 bits (83), Expect = 0.019
Identities = 19/38 (50%), Positives = 22/38 (57%)
Frame = +1
Query: 10 KNVCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQ 47
K V V+GAG SGL A +LK G V VFE GG+
Sbjct: 55 KRVAVVGAGVSGLAAAYKLKSHGLDVTVFEAEGRAGGR 168
>TC78409 homologue to GP|18086418|gb|AAL57665.1 AT3g59050/F17J16_100
{Arabidopsis thaliana}, partial (19%)
Length = 689
Score = 35.8 bits (81), Expect = 0.033
Identities = 17/46 (36%), Positives = 26/46 (55%)
Frame = +1
Query: 2 VSKTNQNSKNVCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQ 47
VS + S +V VIG G +G+ A R L+ +VV+ E +GG+
Sbjct: 376 VSSVQERSPSVIVIGGGMAGIAAARALQDASFQVVLLESRERLGGR 513
>AJ548105
Length = 394
Score = 34.7 bits (78), Expect = 0.074
Identities = 27/100 (27%), Positives = 51/100 (51%), Gaps = 1/100 (1%)
Frame = +1
Query: 290 TFPPSLAPSLSFMGIPSKVLGPHFFESQAKWIAQLLSGKKILPSFEEM-MKSIKELYHSR 348
T PS+ P+++F+G V + E ++ W+A L+ K LPS E+M ++IK++ +
Sbjct: 7 TVHPSI-PNMAFVGYVESVSNLYTSEIRSMWLAGLIDNKFKLPSAEKMHSQTIKDMETMK 183
Query: 349 EVANVPKRYTHSIADNDGFEYFDKYGDNAGFPKLEEWRKE 388
+ KR + G + D+ ++ G+ WRK+
Sbjct: 184 KSTRFYKR---NCITTFGINHNDEICEDLGW---HTWRKK 285
>TC78514 homologue to SP|Q9SMJ3|ZDS_CAPAN Zeta-carotene desaturase
chloroplast precursor (EC 1.14.99.30) (Carotene 7
8-desaturase)., partial (88%)
Length = 2063
Score = 33.1 bits (74), Expect = 0.21
Identities = 15/36 (41%), Positives = 23/36 (63%)
Frame = +3
Query: 12 VCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQ 47
V +IGAG +G+ EL +GH+V ++E IGG+
Sbjct: 396 VAIIGAGLAGMSTAVELLDQGHEVDIYESRTFIGGK 503
>TC86790 similar to PIR|A84861|A84861 probable amine oxidase [imported] -
Arabidopsis thaliana, partial (91%)
Length = 2198
Score = 32.7 bits (73), Expect = 0.28
Identities = 17/46 (36%), Positives = 25/46 (53%)
Frame = +2
Query: 2 VSKTNQNSKNVCVIGAGPSGLVATRELKREGHKVVVFEQNHDIGGQ 47
V + + S +V VIG G +G+ A R L +VV+ E IGG+
Sbjct: 437 VDQQPRRSPSVIVIGGGMAGIAAARALHDASFQVVLLESRDRIGGR 574
>BF631937 similar to GP|10177621|db phytoene dehydrogenase-like {Arabidopsis
thaliana}, partial (26%)
Length = 688
Score = 32.7 bits (73), Expect = 0.28
Identities = 16/33 (48%), Positives = 19/33 (57%)
Frame = +3
Query: 14 VIGAGPSGLVATRELKREGHKVVVFEQNHDIGG 46
+IG G +GL A L R G V V E+ H IGG
Sbjct: 123 IIGGGHNGLTAAAYLARGGLSVAVLERRHIIGG 221
>TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16.90 -
Arabidopsis thaliana, partial (43%)
Length = 1038
Score = 32.7 bits (73), Expect = 0.28
Identities = 25/74 (33%), Positives = 36/74 (47%), Gaps = 16/74 (21%)
Frame = +1
Query: 314 FESQAKWIAQL-LSG-------KKILPSFEEMMKSIKE----LYHSREVANVPKRYTHSI 361
F+S W ++ L+G K IL S + + IKE + HS E+ N+PK H
Sbjct: 568 FDSAVNWFKEMALNGLLPDQKAKNILLSLAKTEEDIKEANELVLHSIEINNLPKVNDHEE 747
Query: 362 ADN----DGFEYFD 371
D+ D +EYFD
Sbjct: 748 DDDDDDEDNYEYFD 789
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,937,789
Number of Sequences: 36976
Number of extensions: 226647
Number of successful extensions: 1098
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1073
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1082
length of query: 433
length of database: 9,014,727
effective HSP length: 99
effective length of query: 334
effective length of database: 5,354,103
effective search space: 1788270402
effective search space used: 1788270402
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0087.18


