

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0086.14
(430 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
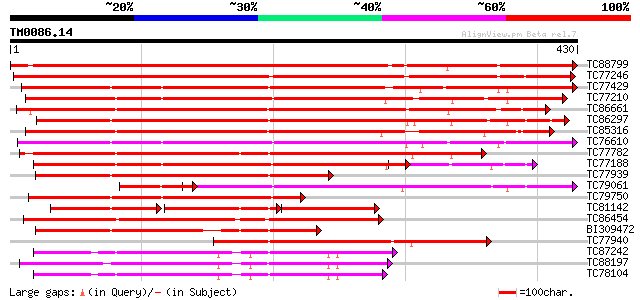
Score E
Sequences producing significant alignments: (bits) Value
TC88799 similar to PIR|C71408|C71408 probable protein kinase - A... 653 0.0
TC77246 similar to PIR|C71408|C71408 probable protein kinase - A... 595 e-170
TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120... 343 6e-95
TC77210 similar to GP|21954717|gb|AAM83095.1 SOS2-like protein k... 338 2e-93
TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine pr... 337 4e-93
TC86297 similar to GP|15912283|gb|AAL08275.1 At1g30270/F12P21_6 ... 333 8e-92
TC85316 weakly similar to PIR|E84707|E84707 probable protein kin... 318 3e-87
TC76610 similar to PIR|T50802|T50802 serine/threonine protein ki... 310 7e-85
TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein k... 300 5e-82
TC77188 similar to PIR|T04862|T04862 probable serine/threonine-s... 289 1e-78
TC77939 similar to GP|14194167|gb|AAK56278.1 At2g26980/T20P8.3 {... 256 2e-68
TC79061 similar to GP|23428550|gb|AAL23677.1 Serine/threonine Ki... 240 9e-64
TC79750 similar to GP|9758929|dbj|BAB09310.1 serine/threonine pr... 239 1e-63
TC81142 similar to GP|8978255|dbj|BAA98146.1 serine/threonine pr... 116 6e-60
TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine... 220 9e-58
BI309472 similar to GP|8978255|dbj serine/threonine protein kina... 209 2e-54
TC77940 similar to PIR|C84667|C84667 probable protein kinase [im... 194 6e-50
TC87242 homologue to PIR|S56716|S56716 protein kinase SPK-3 (EC ... 184 6e-47
TC88197 homologue to GP|6739629|gb|AAF27340.1| abscisic acid-act... 183 1e-46
TC78104 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-ac... 181 5e-46
>TC88799 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (57%)
Length = 1648
Score = 653 bits (1685), Expect = 0.0
Identities = 336/440 (76%), Positives = 375/440 (84%), Gaps = 10/440 (2%)
Frame = +1
Query: 1 MEPPMRQPPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGT-IVAVKIIDKSKT 59
MEPP++ PP PR TILGKYQ+ RLLGRGSFAKVY+A SL+D VAVKIIDKSK
Sbjct: 70 MEPPLQHPPTPRPP---TILGKYQLTRLLGRGSFAKVYKAHSLLDNANTVAVKIIDKSKN 240
Query: 60 VDAAMEPRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKL 119
VDAAMEPRI+REIDAMRRL HHPNIL+IHEVMATKTKIHL+VEFAAGGELFSA+SRRG+
Sbjct: 241 VDAAMEPRIIREIDAMRRLQHHPNILKIHEVMATKTKIHLIVEFAAGGELFSALSRRGRF 420
Query: 120 PENTARRYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGL 179
E+TAR YFQQLVSALRFCHRNGVAHRDLKPQNLLLDA GNLKVSDFGLSALPEHL++GL
Sbjct: 421 SESTARFYFQQLVSALRFCHRNGVAHRDLKPQNLLLDADGNLKVSDFGLSALPEHLKDGL 600
Query: 180 LHTACGTPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISR 239
LHTACGTPAYTAPEILR SGGYDGSKADAWSCGL+L+VLLAGYLPFDD NI AM +KISR
Sbjct: 601 LHTACGTPAYTAPEILRRSGGYDGSKADAWSCGLILYVLLAGYLPFDDLNIPAMCKKISR 780
Query: 240 RDYQFPEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLD 299
RDY+FP+W++K AR VI RLLDPNPETRMS+E L+GN WFKKSLK EA E E S D
Sbjct: 781 RDYRFPDWVSKRARIVIYRLLDPNPETRMSLEGLFGNEWFKKSLKPEA--EPEKSIFGCD 954
Query: 300 SGYGNRVRGLGVNAFDLISMSSGLDLSGLFQ---------DEGKRKEKRFTSGAKLEVVE 350
YG + LG+NAF +ISMSSGLDLSGLF+ + +EKRFTS A +EVV
Sbjct: 955 --YGKEGKNLGLNAFHIISMSSGLDLSGLFETTSSSQGGNNNNTWREKRFTSSANIEVVG 1128
Query: 351 EKVKEIGGVLGFKVEVGKDSTAIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGLEFEE 410
EKVKE+GGVLGFKVEVGK+S AIGLVKGKV LVV+VFEI+P +L+LVAVKVVEGG EFE+
Sbjct: 1129EKVKEVGGVLGFKVEVGKNS-AIGLVKGKVGLVVEVFEIVPCQLVLVAVKVVEGGFEFED 1305
Query: 411 NHWGDWKLGLQDLVLSWYNE 430
NHW DWK+GLQDLV+SW+NE
Sbjct: 1306NHWEDWKVGLQDLVISWHNE 1365
>TC77246 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (65%)
Length = 1715
Score = 595 bits (1533), Expect = e-170
Identities = 300/427 (70%), Positives = 361/427 (84%), Gaps = 1/427 (0%)
Frame = +3
Query: 4 PMRQPPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAA 63
P P PP S + ILGKYQ+ + LGRG+FAKVYQARSLIDG+ VAVK+IDKSKTVDA+
Sbjct: 30 PQSPPSPPPSPPSRIILGKYQLTKFLGRGNFAKVYQARSLIDGSTVAVKVIDKSKTVDAS 209
Query: 64 MEPRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENT 123
MEPRIVREIDAMRRL +HPNIL+IHEV+ATKTKI+L+V+FA GGELF ++RR K PE
Sbjct: 210 MEPRIVREIDAMRRLQNHPNILKIHEVLATKTKIYLIVDFAGGGELFLKLARRRKFPEAL 389
Query: 124 ARRYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTA 183
ARRYFQQLVSAL FCHRNG+AHRDLKPQNLLLDA GNLKVSDFGLSALPE L+NGLLHTA
Sbjct: 390 ARRYFQQLVSALCFCHRNGIAHRDLKPQNLLLDAEGNLKVSDFGLSALPEQLKNGLLHTA 569
Query: 184 CGTPAYTAPEIL-RLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDY 242
CGTPAYTAPEIL R+ GYDGSKADAWSCG++L+VLLAG+LPFDDSNI AM ++I+RRDY
Sbjct: 570 CGTPAYTAPEILGRI--GYDGSKADAWSCGVILYVLLAGHLPFDDSNIPAMCKRITRRDY 743
Query: 243 QFPEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLDSGY 302
QFPEWI+KPAR++I +LLDPNP+TR+ +E+++GN+WFKKSL+ E E + + D
Sbjct: 744 QFPEWISKPARYLIYQLLDPNPKTRIRLENVFGNNWFKKSLREEPEEKLFDDKYSFD--- 914
Query: 303 GNRVRGLGVNAFDLISMSSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEKVKEIGGVLGF 362
G + LG+NAFD+IS+SSGL+LSGLF+ R EKRFTS ++ VVEEKVKEIG LGF
Sbjct: 915 GYKKLELGMNAFDIISLSSGLNLSGLFETTTLRSEKRFTSSEEVGVVEEKVKEIGVRLGF 1094
Query: 363 KVEVGKDSTAIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWGDWKLGLQD 422
+VE+GK+ AIGL KGKV +VV+VF+I+ D+LLLVA+K+ GGLEFE+ HW DWK+GLQD
Sbjct: 1095RVEIGKNG-AIGLGKGKVTMVVEVFKIV-DDLLLVALKLENGGLEFEDVHWNDWKIGLQD 1268
Query: 423 LVLSWYN 429
+VLSW+N
Sbjct: 1269VVLSWHN 1289
>TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120
{Arabidopsis thaliana}, partial (79%)
Length = 1869
Score = 343 bits (881), Expect = 6e-95
Identities = 194/433 (44%), Positives = 274/433 (62%), Gaps = 12/433 (2%)
Frame = +3
Query: 10 PPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIV 69
P A + GKY++ R+LG+G+FAKVY A+ + G VA+K++DK K M +I
Sbjct: 231 PMEEVKAHMLFGKYEMGRVLGKGTFAKVYYAKEISTGEGVAIKVVDKDKVKKEGMMEQIK 410
Query: 70 REIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQ 129
REI MR L HPNI+ + EVMATKTKI V+E+A GGELF +++ GK E AR+YFQ
Sbjct: 411 REISVMR-LVKHPNIVNLKEVMATKTKILFVMEYARGGELFEKVAK-GKFKEELARKYFQ 584
Query: 130 QLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGTPA 188
QL+SA+ +CH GV+HRDLKP+NLLLD NLKVSDFGLSALPEHL Q+GLLHT CGTPA
Sbjct: 585 QLISAVDYCHSRGVSHRDLKPENLLLDENENLKVSDFGLSALPEHLRQDGLLHTQCGTPA 764
Query: 189 YTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWI 248
Y APE++R GY G KAD WSCG++L+ LLAG+LPF N+ +MY K+ + +YQFP W
Sbjct: 765 YVAPEVVR-KRGYSGFKADTWSCGVILYALLAGFLPFQHENLISMYNKVFKEEYQFPPWF 941
Query: 249 TKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLDSGYGNRVRG 308
+ ++ +I ++L +PE R+++ + WF+K L + + ++T N D ++V
Sbjct: 942 SPESKRLISKILVADPERRITISSIMNVPWFQKGLCL-----STTTTANDDFELESKVNL 1106
Query: 309 LG-----VNAFDLI-SMSSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEKVKEIGGVLGF 362
+ NAF+ I SMSSG DLSGLF +E K++ FTS + + K++ L F
Sbjct: 1107INSTPQFFNAFEFISSMSSGFDLSGLF-EEKKKRGSVFTSKCSVSEIVTKIESAAKNLRF 1283
Query: 363 KVEVGKD--STAIGLV---KGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWGDWK 417
KV+ KD GL+ KGK+A+ +++E+ P+ ++ K E+ + D +
Sbjct: 1284KVKKVKDFKLRLQGLIEGRKGKLAVTAEIYEVAPELAVVEFSKCSGDTFEYVKLFEEDVR 1463
Query: 418 LGLQDLVLSWYNE 430
L+D+V SW E
Sbjct: 1464PALKDIVWSWQGE 1502
>TC77210 similar to GP|21954717|gb|AAM83095.1 SOS2-like protein kinase
{Glycine max}, partial (88%)
Length = 1831
Score = 338 bits (867), Expect = 2e-93
Identities = 187/427 (43%), Positives = 272/427 (62%), Gaps = 16/427 (3%)
Frame = +1
Query: 13 SSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREI 72
+S + + GKY++ R+LG G+FAKVY A++L G VA+K++ K K + M +I REI
Sbjct: 325 NSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREI 504
Query: 73 DAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLV 132
M+ + H PNI+++HEVMA+K+KI++ +E GGELF+ I + G+L E+ AR YFQQL+
Sbjct: 505 SVMKMVKH-PNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLI 678
Query: 133 SALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGTPAYTA 191
SA+ FCH GV HRDLKP+NLLLD GNLKVSDFGL EHL Q+GLLHT CGTPAY +
Sbjct: 679 SAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVS 858
Query: 192 PEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKP 251
PE++ G YDG+KAD WSCG++L+VLLAG+LPF D N+ AMY+KI R D++ P W +
Sbjct: 859 PEVIAKKG-YDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLE 1035
Query: 252 ARFVIRRLLDPNPETRMSVEDLYGNSWFKKSL-------KVEAESETESSTLNLDSGYGN 304
AR +I +LLDPNP TR+S+ + +SWFKK + K E E E + ++ +
Sbjct: 1036ARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEFMECEKSS 1215
Query: 305 RVRGLGVNAFDLISMSSGLDLSGLFQDEGK--RKEKRFTSGAKLEVVEEKVKEIGGVLGF 362
+NAF +IS+S G DLS LF+++ + R+E RF +G V K++++ +
Sbjct: 1216TT----MNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAKAV-- 1377
Query: 363 KVEVGKDSTAIGLV------KGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWGDW 416
K +V T + L KGK+A+ ++ + P L++ K LE+ + +
Sbjct: 1378KFDVKSSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKEL 1557
Query: 417 KLGLQDL 423
+ L+D+
Sbjct: 1558RPALKDI 1578
>TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine protein
kinase {Arabidopsis thaliana}, partial (58%)
Length = 1840
Score = 337 bits (865), Expect = 4e-93
Identities = 196/423 (46%), Positives = 271/423 (63%), Gaps = 18/423 (4%)
Frame = +2
Query: 6 RQPPPPRSS------------AATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKI 53
+QPPPPR S A T + GKY++ +LLG G+FAKVY AR+ +G VAVK+
Sbjct: 218 KQPPPPRMSDIEQAAAPAPADATTALFGKYELGKLLGCGAFAKVYHARNTENGQSVAVKV 397
Query: 54 IDKSKTVDAAMEPRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAI 113
I+K K + + REI M +L H PNI+R+HEV+ATKTKI+ V+EFA GGELF+ I
Sbjct: 398 INKKKISATGLAGHVKREISIMSKLRH-PNIVRLHEVLATKTKIYFVMEFAKGGELFAKI 574
Query: 114 SRRGKLPENTARRYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPE 173
+ +G+ E+ +RR FQQL+SA+ +CH +GV HRDLKP+NLLLD GNLKVSDFGLSA+ E
Sbjct: 575 ANKGRFSEDLSRRLFQQLISAVGYCHSHGVFHRDLKPENLLLDDKGNLKVSDFGLSAVKE 754
Query: 174 HLQ-NGLLHTACGTPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAA 232
++ +G+LHT CGTPAY APEIL GYDG+K D WSCG++LFVL+AGYLPF+D N+
Sbjct: 755 QVRIDGMLHTLCGTPAYVAPEIL-AKRGYDGAKVDIWSCGIILFVLVAGYLPFNDPNLMV 931
Query: 233 MYRKISRRDYQFPEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLK-VEAESET 291
MYRKI +++ P W + + + RL+D NPETR++V+++ + WF+K K V+ E
Sbjct: 932 MYRKIYSGEFKCPRWFSPDLKRFLSRLMDTNPETRITVDEILRDPWFRKGYKEVKFYEEG 1111
Query: 292 ESSTLNLDSGYGNRVRGLGVNAFDLIS-MSSGLDLSGLFQD--EGKRKEKRFTSGAKLEV 348
+++G + L NAFD+IS S GL+LSG F D EG+R R +E
Sbjct: 1112FEFEKKVNNG-EEEGKPLDFNAFDIISFFSRGLNLSGFFIDGGEGERFMLRELPEKVVEK 1288
Query: 349 VEEKVKEIGGVLGFKVEVGKDSTAIGLVKGKVALVVQVFEILPDELLLVAVKVVEG-GLE 407
E K G V+ K E G + I G + + ++++ L E+++V VK G +
Sbjct: 1289AEAAAKAEGLVVRRKKECGVE---IEEQNGDLVIGIEIYR-LTAEMVVVEVKRFGGDAVA 1456
Query: 408 FEE 410
FEE
Sbjct: 1457FEE 1465
>TC86297 similar to GP|15912283|gb|AAL08275.1 At1g30270/F12P21_6
{Arabidopsis thaliana}, partial (90%)
Length = 2043
Score = 333 bits (854), Expect = 8e-92
Identities = 185/429 (43%), Positives = 277/429 (64%), Gaps = 25/429 (5%)
Frame = +2
Query: 21 GKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLHH 80
GKY++ R LG G+FAKV AR + G VA+KI+DK K + M +I +EI M+ L
Sbjct: 443 GKYELGRTLGEGNFAKVKFARHIETGDHVAIKILDKEKILKHKMIRQIKQEISTMK-LIR 619
Query: 81 HPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCHR 140
HPN++R+HEV+A ++KI +V+E GGELF I+R G+L E+ AR+YFQQL+ A+ +CH
Sbjct: 620 HPNVIRMHEVIANRSKIFIVMELVTGGELFDKIARSGRLKEDEARKYFQQLICAVDYCHS 799
Query: 141 NGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGTPAYTAPEILRLSG 199
GV HRDLKP+NLLLD G LKVSDFGLSALP+ + ++GLLHT CGTP Y APE+++ +
Sbjct: 800 RGVCHRDLKPENLLLDTNGTLKVSDFGLSALPQQVREDGLLHTTCGTPNYVAPEVIQ-NK 976
Query: 200 GYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRRL 259
GYDG+ AD WSCG++LFVL+AGYLPF++ N+ A+Y+KI + D+ P W + A+ +I+R+
Sbjct: 977 GYDGAIADLWSCGVILFVLMAGYLPFEEDNLVALYKKIFKADFTCPPWFSSSAKKLIKRI 1156
Query: 260 LDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLDS---GYGNR----------- 305
LDP+P TR+ + ++ N WFKK K + + S +++S GY +
Sbjct: 1157LDPSPITRIKIAEVIENEWFKKGYKPPVFEQADVSLDDVNSIFNGYMDSDNIVVERQEEG 1336
Query: 306 -VRGLGVNAFDLISMSSGLDLSGLFQDEG--KRKEKRFTSGAKLEVVEEKVKEIGGVLGF 362
V + +NAF+LIS S GL+LS LF+ + ++E RFTS + K+++ G LGF
Sbjct: 1337PVAPVTMNAFELISKSQGLNLSSLFEKQMGLVKRETRFTSKCSANEIISKIEKTAGPLGF 1516
Query: 363 KVEVGKDSTAIGLV------KGKVALVVQVFEILPDELLLVAVKVVEGG-LEFEENHWGD 415
+V K++ + + KG +++ ++ E+ P + +V ++ EG LEF + +
Sbjct: 1517--DVKKNNCKLRIYGEKTGRKGHLSVATEILEVAP-SIYMVEMRKSEGDTLEFHK-FYKS 1684
Query: 416 WKLGLQDLV 424
GL+D+V
Sbjct: 1685ITTGLKDIV 1711
>TC85316 weakly similar to PIR|E84707|E84707 probable protein kinase
[imported] - Arabidopsis thaliana, partial (66%)
Length = 1528
Score = 318 bits (815), Expect = 3e-87
Identities = 182/410 (44%), Positives = 266/410 (64%), Gaps = 9/410 (2%)
Frame = +1
Query: 13 SSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVD-AAMEPRIVRE 71
S+ + + GKY++ RLLG G+ AKVY A ++ G VAVK+I K K ++ I RE
Sbjct: 232 STNSVVLFGKYEIGRLLGVGASAKVYHATNIETGKSVAVKVISKKKLINNGEFAANIERE 411
Query: 72 IDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQL 131
I +RRLHH PNI+ + EV+A+K+KI+ VVEFA GELF ++++ KL E+ ARRYF+QL
Sbjct: 412 ISILRRLHH-PNIIDLFEVLASKSKIYFVVEFAEAGELFEEVAKKEKLTEDHARRYFRQL 588
Query: 132 VSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQ-NGLLHTACGTPAYT 190
+SA++ CH GV HRDLK NLLLD NLKV+DFGLSA+ ++ +GLLHT CGTP+Y
Sbjct: 589 ISAVKHCHSRGVFHRDLKLDNLLLDENDNLKVTDFGLSAVKNQIRPDGLLHTVCGTPSYV 768
Query: 191 APEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITK 250
APEIL GY+G+KAD WSCG+VLF + AGYLPF+D N+ +YRKI R ++FP+W++
Sbjct: 769 APEIL-AKKGYEGAKADVWSCGVVLFTVTAGYLPFNDYNVTVLYRKIYRGQFRFPKWMSC 945
Query: 251 PARFVIRRLLDPNPETRMSVEDLYGNSWFK----KSLKVEAESETESSTLNLDSGYGNRV 306
+ ++ R+LD NP+TR+SV+++ + WF K +V +S+ E S S
Sbjct: 946 DLKNLLSRMLDTNPKTRISVDEILEDPWFSSGGYKLDRVLVKSQMEESRTGFKS------ 1107
Query: 307 RGLGVNAFDLISMSSGLDLSGLFQDE-GKRKEKRFTSGAKLEVVEEKVKEIGG--VLGFK 363
+NAFDLIS S+GLD+SG+F++ G +R S E + ++V+E+ G V+ +
Sbjct: 1108----LNAFDLISFSTGLDMSGMFEENVGLSLVERVVSAKLPERIVKRVEEVVGTKVVVKR 1275
Query: 364 VEVGKDSTAIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHW 413
++ G + G +ALVV +++ L +EL++V +K G E+ W
Sbjct: 1276LKNGGGARLEGQEGNLIALVV-IYQ-LTEELVVVEMKKRGKGDEYNAQLW 1419
>TC76610 similar to PIR|T50802|T50802 serine/threonine protein kinase-like
protein - Arabidopsis thaliana, partial (73%)
Length = 1799
Score = 310 bits (794), Expect = 7e-85
Identities = 191/453 (42%), Positives = 266/453 (58%), Gaps = 29/453 (6%)
Frame = +2
Query: 7 QPPPPRSSAATTIL-GKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAME 65
QP P IL KY++ + LG+G+FAKVY R++ VA+K+I K K +
Sbjct: 275 QPLPMEQHRKRNILFNKYEIGKTLGQGNFAKVYHGRNIATNENVAIKVIKKEKLKKERLM 454
Query: 66 PRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTAR 125
+I RE+ MR L HP+I+ + EVMA K K+ +VVE+ GGELF+ +++ GK+ EN AR
Sbjct: 455 KQIKREVSVMR-LVRHPHIVELKEVMANKAKVFMVVEYVKGGELFAKVAK-GKMEENIAR 628
Query: 126 RYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQN-GLLHTAC 184
+YFQQL+SA+ FCH GV HRDLKP+NLLLD +LKVSDFGLSALP+ ++ G+L T C
Sbjct: 629 KYFQQLISAVDFCHSRGVTHRDLKPENLLLDENEDLKVSDFGLSALPDQRRSDGMLLTPC 808
Query: 185 GTPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQF 244
GTPAY APE+L+ G YDGSKAD WSCG++LF LL GYLPF N+ +Y K + DY
Sbjct: 809 GTPAYVAPEVLKKIG-YDGSKADIWSCGVILFALLCGYLPFQGENVMRIYSKSFKADYVL 985
Query: 245 PEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKS-LKVEAESETESST-LNLD--- 299
PEWI+ A+ +I LL +PE R S+ D+ + WF+ ++ A S ES+ N+D
Sbjct: 986 PEWISPGAKKLISNLLVVDPEKRFSIPDIMKDPWFQIGFMRPIAFSMKESAVDDNIDFSG 1165
Query: 300 ----SGYGNRVRGLGV-----------NAFDLI-SMSSGLDLSGLFQDEGKRKEKRFTSG 343
+G GN V +GV NAF++I S+S G DL LF+ KR F S
Sbjct: 1166DDEGNGDGNSVEVVGVTGTKHSRRPSYNAFEIISSLSHGFDLRNLFETR-KRSPSMFISK 1342
Query: 344 AKLEVVEEKVKEIGGVLGFKVEVGKD------STAIGLVKGKVALVVQVFEILPDELLLV 397
V K++ + L +V KD T G KGK+ + V+VFE+ P+ ++
Sbjct: 1343LSASAVVGKLENVAKKLNLRVTGKKDFMVRMQGTTEGR-KGKLGMTVEVFEVAPEVAVVE 1519
Query: 398 AVKVVEGGLEFEENHWGDWKLGLQDLVLSWYNE 430
K LE+ + + + L+D+V SW +
Sbjct: 1520FSKSAGDTLEYVKFCEDEVRPSLKDIVWSWQGD 1618
>TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein kinase PKS6
{Arabidopsis thaliana}, partial (77%)
Length = 1236
Score = 300 bits (769), Expect = 5e-82
Identities = 160/366 (43%), Positives = 235/366 (63%), Gaps = 12/366 (3%)
Frame = +2
Query: 8 PPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPR 67
PP PR T +GKY++ + +G GSFAKV A+++ +G VA+KI+D++ + M +
Sbjct: 161 PPRPR-----TRVGKYEMGKTIGEGSFAKVKLAKNVENGDYVAIKILDRNHVLKHNMMDQ 325
Query: 68 IVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRY 127
+ REI AM+ ++H PN+++I EVMA+KTKI++V+E GGELF I+ G+L E+ AR Y
Sbjct: 326 LKREISAMKIINH-PNVIKIFEVMASKTKIYIVLELVNGGELFDKIATHGRLKEDEARSY 502
Query: 128 FQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTP 187
FQQL++A+ +CH GV HRDLKP+NLLLD G LKVSDFGLS + ++ LL TACGTP
Sbjct: 503 FQQLINAVDYCHSRGVYHRDLKPENLLLDTNGVLKVSDFGLSTYSQQ-EDELLRTACGTP 679
Query: 188 AYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEW 247
Y APE+L GY GS +D WSCG++LFVL+AGYLPFD+ N+ A+YRKI R D++ P W
Sbjct: 680 NYVAPEVLN-DRGYVGSSSDIWSCGVILFVLMAGYLPFDEPNLIALYRKIGRADFKCPSW 856
Query: 248 ITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLDSGYGN--- 304
+ A+ ++ +L+PNP TR+ + ++ + WF+K K +E E ++ + N
Sbjct: 857 FSPEAKKLLTSILNPNPLTRIKIPEILEDEWFRKGYKPACFTEEEDVNVDDVAAAFNDSR 1036
Query: 305 -------RVRGLGVNAFDLISMSSGLDLSGLFQDEG--KRKEKRFTSGAKLEVVEEKVKE 355
+ + + +NAF+LIS S +L LF+ E ++E FTS + K++E
Sbjct: 1037ENLVTETKEKPVSMNAFELISRSESFNLDNLFEREKGVVKRETHFTSQRPANEIMSKIEE 1216
Query: 356 IGGVLG 361
LG
Sbjct: 1217AAKPLG 1234
>TC77188 similar to PIR|T04862|T04862 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) F28A21.110 - Arabidopsis
thaliana, partial (79%)
Length = 1881
Score = 289 bits (740), Expect = 1e-78
Identities = 149/293 (50%), Positives = 207/293 (69%), Gaps = 7/293 (2%)
Frame = +1
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRL 78
+LG++++ +LLG G+FAKV+ A++L G VA+KII K K + + + I REI +RR+
Sbjct: 178 LLGRFELGKLLGHGTFAKVHLAKNLKTGESVAIKIISKDKILKSGLVSHIKREISILRRV 357
Query: 79 HHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFC 138
H PNI+++ EVMATKTKI+ V+E+ GGELF+ +++ G+L E AR+YFQQL+ A+ FC
Sbjct: 358 RH-PNIVQLFEVMATKTKIYFVMEYVRGGELFNKVAK-GRLKEEVARKYFQQLICAVGFC 531
Query: 139 HRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGTPAYTAPEILRL 197
H GV HRDLKP+NLLLD GNLKVSDFGLSA+ + + Q+GL HT CGTPAY APE+L
Sbjct: 532 HARGVFHRDLKPENLLLDEKGNLKVSDFGLSAVSDEIKQDGLFHTFCGTPAYVAPEVLSR 711
Query: 198 SGGYDGSKADAWSCGLVLFVLLAGYLPF-DDSNIAAMYRKISRRDYQFPEWITKPARFVI 256
GYDG+K D WSCG+VLFVL+AGYLPF D +N+ AMY+KI + +++ P W + ++
Sbjct: 712 K-GYDGAKVDIWSCGVVLFVLMAGYLPFHDPNNVMAMYKKIYKGEFRCPRWFSPELVSLL 888
Query: 257 RRLLDPNPETRMSVEDLYGNSWFKKSLK-----VEAESETESSTLNLDSGYGN 304
RLLD P+TR+S+ ++ N WFK K VE + + +L+LD N
Sbjct: 889 TRLLDIKPQTRISIPEIMENRWFKIGFKHIKFYVEDDVVCDLDSLDLDGEDNN 1047
Score = 63.9 bits (154), Expect = 1e-10
Identities = 42/119 (35%), Positives = 70/119 (58%), Gaps = 6/119 (5%)
Frame = +2
Query: 288 ESETESSTLNLD-SGYGNRVRGLGVNAFDLISMSSGLDLSGLFQDEGKRKEKRFTSGAKL 346
++E++S +N G+ R +NAFD+IS S G DLSGLF+++G E RF S A +
Sbjct: 1103 KNESDSEVVNRRIRNRGSLPRPASLNAFDIISFSQGFDLSGLFEEKG--DEARFVSSASV 1276
Query: 347 EVVEEKVKEIGGVLGFKV-----EVGKDSTAIGLVKGKVALVVQVFEILPDELLLVAVK 400
+ K++E+G ++ F V +V + + G KG++ + +VFE+ P L++V VK
Sbjct: 1277 PKIISKLEEVGQMVRFNVRKKDCKVSLEGSREG-AKGRLTIAAEVFELTP-SLVVVEVK 1447
>TC77939 similar to GP|14194167|gb|AAK56278.1 At2g26980/T20P8.3 {Arabidopsis
thaliana}, partial (70%)
Length = 1353
Score = 256 bits (653), Expect = 2e-68
Identities = 128/227 (56%), Positives = 167/227 (73%), Gaps = 1/227 (0%)
Frame = +1
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G Y+V R +G G+FAKV AR+ G VA+KI+DK K + M +I REI M+ L
Sbjct: 544 VGIYEVGRTIGEGTFAKVKFARNSETGEAVALKILDKEKVLKHKMAEQIKREIATMK-LI 720
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
HPN++R++EVM ++TKI++V+EF GGELF I G++ E ARRYFQQL++ + +CH
Sbjct: 721 KHPNVVRLYEVMGSRTKIYIVLEFVTGGELFDKIVNHGRMGEPEARRYFQQLINVVDYCH 900
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQ-NGLLHTACGTPAYTAPEILRLS 198
GV HRDLKP+NLLLDA GNLKVSDFGLSAL + ++ +GLLHT CGTP Y APE+L
Sbjct: 901 SRGVYHRDLKPENLLLDAYGNLKVSDFGLSALSQQVRDDGLLHTTCGTPNYVAPEVLN-D 1077
Query: 199 GGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFP 245
GYDG+ AD WSCG++LFVL+AGYLPFDD N+ +Y+KIS D+ P
Sbjct: 1078RGYDGATADLWSCGVILFVLVAGYLPFDDPNLMELYKKISSADFTCP 1218
>TC79061 similar to GP|23428550|gb|AAL23677.1 Serine/threonine Kinase
{Persea americana}, partial (72%)
Length = 1367
Score = 240 bits (612), Expect = 9e-64
Identities = 140/325 (43%), Positives = 195/325 (59%), Gaps = 26/325 (8%)
Frame = +2
Query: 132 VSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGTPAYT 190
++ L +CH GV HRDLKP+NLLLD NLKVSDFGLS L E Q+GLLHT CGTPAY
Sbjct: 143 LAQLTYCHSRGVYHRDLKPENLLLDENENLKVSDFGLSTLSESKSQDGLLHTTCGTPAYV 322
Query: 191 APEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITK 250
APE++ G Y+G KAD WSCG++L+VLLAGYLPF D N+ MYRKI + +++FP+W
Sbjct: 323 APEVINRKG-YEGCKADIWSCGVILYVLLAGYLPFHDQNLMEMYRKIGKGEFKFPKWFAP 499
Query: 251 PARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTL-------------- 296
R ++ ++LDP+ +TR+S+ + NSWFKK L+ ETE++ L
Sbjct: 500 EVRRLLSKILDPSLKTRISMAKIMENSWFKKGLEKPVVIETENNELTSLHAEGVFEVSEN 679
Query: 297 ----NLDSGYGNRVRGLGVNAFDLISMSSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEK 352
N ++ +NAFD+IS SSG DLSGLF+D ++KE RFTS + K
Sbjct: 680 GGDSNTETKQLQAKPCNNLNAFDIISFSSGFDLSGLFEDTIQKKEMRFTSNKPASTIISK 859
Query: 353 VKEIGGVLGFKVEVGKDSTAIGLV------KGKVALVVQVFEILPDELLLVAVKVVEGG- 405
++EI L KV+ KD + L KG + + ++FEI P LV +K +G
Sbjct: 860 LEEICKCLQLKVK-KKDGGLLKLEGSKEGRKGILNIDAEIFEITP-HFHLVELKKSDGDT 1033
Query: 406 LEFEENHWGDWKLGLQDLVLSWYNE 430
+E+E+ + + L+D+V +W E
Sbjct: 1034VEYEKLLQQEIRPALKDIVWNWQGE 1108
Score = 63.5 bits (153), Expect = 1e-10
Identities = 32/59 (54%), Positives = 45/59 (76%)
Frame = +1
Query: 84 ILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCHRNG 142
++ ++EVMA+KTKI +V+E+A GGELF I+ +GKL + ARRYFQQLVSA+ + G
Sbjct: 1 VVELYEVMASKTKIFIVMEYARGGELFHKIA-KGKLKTDVARRYFQQLVSAVDLLPQQG 174
>TC79750 similar to GP|9758929|dbj|BAB09310.1 serine/threonine protein
kinase {Arabidopsis thaliana}, partial (47%)
Length = 1101
Score = 239 bits (610), Expect = 1e-63
Identities = 120/211 (56%), Positives = 162/211 (75%), Gaps = 1/211 (0%)
Frame = +2
Query: 15 AATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDA 74
+ + I+ +Y++ +LLG+G+FAKVY R+L + VA+K+IDK K + M +I REI
Sbjct: 470 SGSVIMQRYELGKLLGQGTFAKVYHGRNLKNSMSVAIKVIDKEKVLKVGMIDQIKREISV 649
Query: 75 MRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSA 134
MR L HP+++ ++EVMA+KTKI++V+E+ GGELF+ +S+ GKL + ARRYFQQL+SA
Sbjct: 650 MR-LIRHPHVVELYEVMASKTKIYVVMEYVKGGELFNKVSK-GKLKHDEARRYFQQLISA 823
Query: 135 LRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPE-HLQNGLLHTACGTPAYTAPE 193
+ +CH GV HRDLKP+NLLLD GNLKVSDFGLSAL E Q+GLLHT CGTPAY APE
Sbjct: 824 VDYCHSRGVCHRDLKPENLLLDENGNLKVSDFGLSALAETKHQDGLLHTTCGTPAYVAPE 1003
Query: 194 ILRLSGGYDGSKADAWSCGLVLFVLLAGYLP 224
++ GYDG+KAD W+ G++L+VLLAG+LP
Sbjct: 1004VINRK-GYDGTKADIWAWGVILYVLLAGFLP 1093
>TC81142 similar to GP|8978255|dbj|BAA98146.1 serine/threonine protein
kinase SOS2 {Arabidopsis thaliana}, partial (54%)
Length = 784
Score = 116 bits (291), Expect(3) = 6e-60
Identities = 59/89 (66%), Positives = 69/89 (77%)
Frame = +1
Query: 118 KLPENTARRYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQN 177
KL EN +RRYFQQL+ A+ CH+ GV HRDLKP+NLLLDA GNLKVSDFGLSAL + +
Sbjct: 301 KLSENESRRYFQQLIDAVAHCHKKGVYHRDLKPENLLLDAYGNLKVSDFGLSALTKQ-GD 477
Query: 178 GLLHTACGTPAYTAPEILRLSGGYDGSKA 206
LLHT CGTP Y APE+L + GYDG+ A
Sbjct: 478 ELLHTTCGTPNYVAPEVLS-NQGYDGAAA 561
Score = 81.3 bits (199), Expect(3) = 6e-60
Identities = 29/74 (39%), Positives = 55/74 (74%)
Frame = +2
Query: 207 DAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRRLLDPNPET 266
D WSCG++L+VL+AGYLPF+++++ ++R+IS ++ P W + A+ I ++LDP+P+T
Sbjct: 563 DVWSCGIILYVLMAGYLPFEEADLPTLFRRISAGEFVCPVWFSAGAKTFIHKILDPDPKT 742
Query: 267 RMSVEDLYGNSWFK 280
R+ + ++ + WF+
Sbjct: 743 RVKIVEIRKDPWFR 784
Score = 72.4 bits (176), Expect(3) = 6e-60
Identities = 37/84 (44%), Positives = 57/84 (67%)
Frame = +3
Query: 32 GSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLHHHPNILRIHEVM 91
G+FAKV A+ G VA+K++ K+ + M +I REI M+ + HPNI+R+HEV+
Sbjct: 6 GTFAKVKYAKHSETGESVAIKVMAKTTILKHRMVEQIKREISIMK-IVRHPNIVRLHEVL 182
Query: 92 ATKTKIHLVVEFAAGGELFSAISR 115
A++TKI++++EF GGEL+ I R
Sbjct: 183 ASQTKIYIILEFVMGGELYDKIVR 254
>TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine/threonine
protein kinase {Glycine max}, partial (90%)
Length = 2194
Score = 220 bits (560), Expect = 9e-58
Identities = 116/275 (42%), Positives = 177/275 (64%), Gaps = 2/275 (0%)
Frame = +3
Query: 11 PRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVR 70
P L Y++ + LG GSF KV A ++ G VA+KI+++ K + ME ++ R
Sbjct: 78 PGGGNVNAFLRNYKMGKTLGIGSFGKVKIAEHVLTGHKVAIKILNRRKIKNMEMEEKVRR 257
Query: 71 EIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQ 130
EI +R HH +I+R++EV+ T T I++V+E+ GELF I +G+L E+ AR +FQQ
Sbjct: 258 EIKILRLFMHH-HIIRLYEVVETPTDIYVVMEYVKSGELFDYIVEKGRLQEDEARSFFQQ 434
Query: 131 LVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNG-LLHTACGTPAY 189
++S + +CHRN V HRDLKP+NLLLD+ ++K++DFGLS + +++G L T+CG+P Y
Sbjct: 435 IISGVEYCHRNMVVHRDLKPENLLLDSKWSVKIADFGLSNI---MRDGHFLKTSCGSPNY 605
Query: 190 TAPEILRLSGG-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWI 248
APE+ +SG Y G + D WSCG++L+ LL G LPFDD NI +++KI Y P +
Sbjct: 606 AAPEV--ISGKLYAGPEVDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGIYTLPSHL 779
Query: 249 TKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSL 283
+ AR +I RLL +P RM++ ++ + WF+ L
Sbjct: 780 SPGARDLIPRLLVVDPMKRMTIPEIRQHPWFQLHL 884
>BI309472 similar to GP|8978255|dbj serine/threonine protein kinase SOS2
{Arabidopsis thaliana}, partial (46%)
Length = 726
Score = 209 bits (531), Expect = 2e-54
Identities = 106/217 (48%), Positives = 152/217 (69%)
Frame = +2
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+GKY+V R +G G+FAKV A+ G VA+K++ K+ + M +I REI M+ +
Sbjct: 122 IGKYEVGRTIGEGTFAKVKYAKHSETGESVAIKVMAKTTILKHRMVEQIKREISIMK-IV 298
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
HPNI+R+HEV+A++TKI++++EF GGEL+ I ++ KL EN +RRYFQQL+ A+ CH
Sbjct: 299 RHPNIVRLHEVLASQTKIYIILEFVMGGELYDKIVQQVKLSENESRRYFQQLIDAVAHCH 478
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
+ GV HRD LKVSDFGLSAL + + LLHT CGTP Y APE+L +
Sbjct: 479 KKGVYHRD-------------LKVSDFGLSALTKQ-GDELLHTTCGTPNYVAPEVLS-NQ 613
Query: 200 GYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRK 236
GYDG+ AD WSCG++L+VL+AGYLPF++++++ ++RK
Sbjct: 614 GYDGAAADVWSCGIILYVLMAGYLPFEEADLSTLFRK 724
>TC77940 similar to PIR|C84667|C84667 probable protein kinase [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1258
Score = 194 bits (493), Expect = 6e-50
Identities = 106/226 (46%), Positives = 143/226 (62%), Gaps = 15/226 (6%)
Frame = +2
Query: 155 LDAAGNLKVSDFGLSALPEHLQN-GLLHTACGTPAYTAPEILRLSGGYDGSKADAWSCGL 213
LDA GNLKVSDFGLSAL + +++ GLLHT CGTP Y APE+L GYDG+ AD WSCG+
Sbjct: 2 LDAYGNLKVSDFGLSALSQQVRDDGLLHTTCGTPNYVAPEVLN-DRGYDGATADLWSCGV 178
Query: 214 VLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRRLLDPNPETRMSVEDL 273
+LFVL+AGYLPFDD N+ +Y+KIS D+ P W++ AR +I R+LDPNP TR+++ ++
Sbjct: 179 ILFVLVAGYLPFDDPNLMELYKKISSADFTCPPWLSFSARKLITRILDPNPMTRITMAEI 358
Query: 274 YGNSWFKKSLKVEAESETESSTLNLDSGYG-------------NRVRGLGVNAFDLISMS 320
+ WFKK K ES NLD + +NAF+LISMS
Sbjct: 359 LDDEWFKKDYKPPVFE--ESGETNLDDVEAVFKDSEEHHVTEKKEEQPTSMNAFELISMS 532
Query: 321 SGLDLSGLFQ-DEGKRKEKRFTSGAKLEVVEEKVKEIGGVLGFKVE 365
GL+L LF ++G ++E RFTS + + + K++E LGF V+
Sbjct: 533 RGLNLENLFDVEQGFKRETRFTSQSPADEIINKIEEAAKPLGFDVQ 670
>TC87242 homologue to PIR|S56716|S56716 protein kinase SPK-3 (EC 2.7.1.-) -
soybean, complete
Length = 1659
Score = 184 bits (467), Expect = 6e-47
Identities = 107/288 (37%), Positives = 167/288 (57%), Gaps = 12/288 (4%)
Frame = +1
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRL 78
++ KY++++ LG G+F R+ +VA+K I++ + +D + REI R L
Sbjct: 466 MMDKYELVKDLGAGNFGVARLLRNKETKELVAMKYIERGQKIDE----NVAREIINHRSL 633
Query: 79 HHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFC 138
H PNI+R EV+ T T + +V+E+AAGGELF I G+ E+ AR +FQQL+S + +C
Sbjct: 634 RH-PNIIRFKEVVLTPTHLAIVMEYAAGGELFERICNAGRFSEDEARYFFQQLISGVHYC 810
Query: 139 HRNGVAHRDLKPQNLLLDA--AGNLKVSDFGLSALPEHLQNGLLH----TACGTPAYTAP 192
H + HRDLK +N LLD A LK+ DFG S ++ LLH + GTPAY AP
Sbjct: 811 HAMQICHRDLKLENTLLDGSPAPRLKICDFGYS------KSSLLHSRPKSTVGTPAYIAP 972
Query: 193 EILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRR----DYQFPEW- 247
E+L YDG AD WSCG+ L+V+L G PF+D + +RK +R Y+ P++
Sbjct: 973 EVLS-RREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPRNFRKTIQRIMAVQYKIPDYV 1149
Query: 248 -ITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESS 294
I++ + ++ R+ NP R+S++++ + WF K+L E +++
Sbjct: 1150HISQDCKHLLSRIFVANPLRRISLKEIKNHPWFLKNLPRELTESAQAA 1293
>TC88197 homologue to GP|6739629|gb|AAF27340.1| abscisic acid-activated
protein kinase {Vicia faba}, partial (96%)
Length = 1158
Score = 183 bits (465), Expect = 1e-46
Identities = 110/295 (37%), Positives = 163/295 (54%), Gaps = 12/295 (4%)
Frame = +1
Query: 8 PPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPR 67
PPP +Y +R +G G+F R +VAVK I++ +D ++
Sbjct: 82 PPPAMDMPIMHDSDRYDFVRDIGSGNFGVARLMRDKHTKELVAVKYIERGDKIDENVKRE 261
Query: 68 IVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRY 127
I+ R HPNI+R EV+ T T + +V+E+A+GGE+F ISR G+ E+ AR +
Sbjct: 262 IINH-----RSLRHPNIVRFKEVILTPTHLAIVMEYASGGEMFDRISRAGRFTEDEARFF 426
Query: 128 FQQLVSALRFCHRNGVAHRDLKPQNLLLDA--AGNLKVSDFGLSALPEHLQNGLLH---- 181
FQQL+S + +CH V HRDLK +N LLD A +LK+ DFG S ++ +LH
Sbjct: 427 FQQLISGVSYCHSMQVCHRDLKLENTLLDGDPALHLKICDFGYS------KSSVLHSQPK 588
Query: 182 TACGTPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRR- 240
+ GTPAY APE+L L YDG AD WSCG+ L+V+L G PF+D + +RK +R
Sbjct: 589 STVGTPAYIAPEVL-LKQEYDGKVADVWSCGVTLYVMLVGSYPFEDPDNPKDFRKTIQRV 765
Query: 241 ---DYQFPEW--ITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESE 290
Y P++ I+ R VI R+ +P R+++ ++ N WF K+L + +E
Sbjct: 766 LSVQYSVPDFVQISPECREVISRIFVFDPAKRITIPEILKNEWFLKNLPADLVNE 930
>TC78104 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-activated
protein kinase {Nicotiana tabacum}, partial (95%)
Length = 1434
Score = 181 bits (459), Expect = 5e-46
Identities = 106/280 (37%), Positives = 161/280 (56%), Gaps = 12/280 (4%)
Frame = +3
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRL 78
++ KY+V++ +G G+F R +VA+K I++ +D + REI R L
Sbjct: 150 VMEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDE----NVAREIINHRSL 317
Query: 79 HHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFC 138
H PNI+R EV+ T T + +V+E+AAGGELF I G+ E+ AR +FQQL+S + +C
Sbjct: 318 RH-PNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYC 494
Query: 139 HRNGVAHRDLKPQNLLLDA--AGNLKVSDFGLSALPEHLQNGLLH----TACGTPAYTAP 192
H + HRDLK +N LLD A LK+ DFG S ++ LLH + GTPAY AP
Sbjct: 495 HSMQICHRDLKLENTLLDGSPAPRLKICDFGYS------KSSLLHSRPKSTVGTPAYIAP 656
Query: 193 EILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRR----DYQFPEW- 247
E+L YDG AD WSCG+ L+V+L G PF+D + +RK R Y+ P++
Sbjct: 657 EVLS-RREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYV 833
Query: 248 -ITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVE 286
I++ R ++ R+ +P R++++++ + WF K+L E
Sbjct: 834 HISQECRHLLSRIFVASPARRITIKEIKSHPWFLKNLPRE 953
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,758,038
Number of Sequences: 36976
Number of extensions: 174652
Number of successful extensions: 1969
Number of sequences better than 10.0: 492
Number of HSP's better than 10.0 without gapping: 1620
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1730
length of query: 430
length of database: 9,014,727
effective HSP length: 99
effective length of query: 331
effective length of database: 5,354,103
effective search space: 1772208093
effective search space used: 1772208093
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0086.14


