

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.2
(153 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
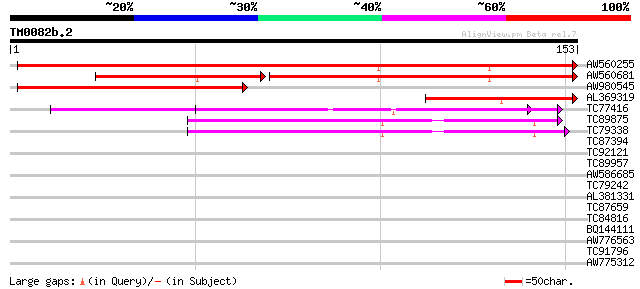
Score E
Sequences producing significant alignments: (bits) Value
AW560255 weakly similar to GP|10086260|gb| calmodulin-binding pr... 173 2e-44
AW560681 weakly similar to GP|10086260|gb| calmodulin-binding pr... 94 2e-30
AW980545 weakly similar to PIR|T10654|T106 hypothetical protein ... 103 2e-23
AL369319 weakly similar to PIR|T10654|T106 hypothetical protein ... 69 7e-13
TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl g... 50 4e-07
TC89875 similar to GP|20259245|gb|AAM14358.1 putative N-terminal... 42 7e-05
TC79338 homologue to GP|20259245|gb|AAM14358.1 putative N-termin... 42 7e-05
TC87394 similar to GP|3582779|gb|AAC69180.1| peroxisomal targeti... 34 0.026
TC92121 weakly similar to GP|10177302|dbj|BAB10563. gene_id:MDC1... 31 0.17
TC89957 weakly similar to PIR|D96810|D96810 hypothetical protein... 30 0.50
AW586685 similar to PIR|T01081|T01 hypothetical protein T10P11.3... 28 1.4
TC79242 26 5.5
AL381331 weakly similar to GP|14209525|dbj contains ESTs C73715(... 26 7.2
TC87659 similar to GP|10177021|dbj|BAB10259. contains similarity... 25 9.3
TC84816 similar to GP|2313789|gb|AAD07729.1| H. pylori predicted... 25 9.3
BQ144111 25 9.3
AW776563 similar to GP|21304447|em HOBBIT protein {Arabidopsis t... 25 9.3
TC91796 GP|23498903|emb|CAD50981. hypothetical protein {Plasmodi... 25 9.3
AW775312 similar to GP|20466732|gb unknown protein {Arabidopsis ... 25 9.3
>AW560255 weakly similar to GP|10086260|gb| calmodulin-binding protein MPCBP
{Zea mays}, partial (7%)
Length = 610
Score = 173 bits (439), Expect = 2e-44
Identities = 86/154 (55%), Positives = 115/154 (73%), Gaps = 3/154 (1%)
Frame = -2
Query: 3 RDYARNLEEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYK 62
R+ R LE E+WHDLA Y +LS+WHDAE+CL+KS+A Y+ASR H G ++EA+GL++
Sbjct: 522 RNRDRRLEVEVWHDLANVYTALSRWHDAEICLAKSQAIDPYSASRLHSTGLLNEARGLHQ 343
Query: 63 EAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDP-VVRSFLMDALRYDRLNASAWYNL 121
EA+K+++ AL+I+P HV SLISTA LR+ QS +VRS L DAL+ D N+SAWYNL
Sbjct: 342 EALKSYKKALDIEPNHVASLISTACVLRKLGGQSSSLIVRSLLTDALKLDTTNSSAWYNL 163
Query: 122 GILHKAE--GRVLEAVECFQAANSLEETAPIEPF 153
G+L+KA+ LEA ECF+ A LEE++PIEPF
Sbjct: 162 GLLYKADLGTSALEAAECFETAVFLEESSPIEPF 61
>AW560681 weakly similar to GP|10086260|gb| calmodulin-binding protein MPCBP
{Zea mays}, partial (3%)
Length = 551
Score = 93.6 bits (231), Expect(2) = 2e-30
Identities = 50/86 (58%), Positives = 62/86 (71%), Gaps = 3/86 (3%)
Frame = -2
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDP-VVRSFLMDALRYDRLNASAWYNLGILHKAE- 128
AL+I+P HV SLISTA LR+ QS +VRS L DAL+ D N+SAWYNLG+L+KA+
Sbjct: 400 ALDIEPNHVASLISTACVLRKLGGQSSSLIVRSLLTDALKLDTTNSSAWYNLGLLYKADL 221
Query: 129 -GRVLEAVECFQAANSLEETAPIEPF 153
LEA ECF+ A LEE++PIEPF
Sbjct: 220 GTSALEAAECFETAVFLEESSPIEPF 143
Score = 54.7 bits (130), Expect(2) = 2e-30
Identities = 24/49 (48%), Positives = 38/49 (76%), Gaps = 3/49 (6%)
Frame = -3
Query: 24 LSQWHDAEVCLSKSKAFRQYTASRCH---VIGTMHEAKGLYKEAVKAFR 69
LS+WHDAE+CL+KS+A Y+A R H G ++EA+GL++EA+K+++
Sbjct: 549 LSRWHDAEICLAKSQAIDPYSAYRLHSTVYAGLLNEARGLHQEALKSYK 403
>AW980545 weakly similar to PIR|T10654|T106 hypothetical protein T5F17.50 -
Arabidopsis thaliana, partial (11%)
Length = 562
Score = 103 bits (258), Expect = 2e-23
Identities = 46/62 (74%), Positives = 53/62 (85%)
Frame = +2
Query: 3 RDYARNLEEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYK 62
RD ARNLE EIWHDLA+ YISLSQWHDA CLSKSKA + Y+ASRCH +G M+EAKGL+K
Sbjct: 377 RDRARNLEVEIWHDLAHVYISLSQWHDAHACLSKSKAIKPYSASRCHALGIMYEAKGLFK 556
Query: 63 EA 64
E+
Sbjct: 557 ES 562
>AL369319 weakly similar to PIR|T10654|T106 hypothetical protein T5F17.50 -
Arabidopsis thaliana, partial (6%)
Length = 318
Score = 68.9 bits (167), Expect = 7e-13
Identities = 31/44 (70%), Positives = 38/44 (85%), Gaps = 3/44 (6%)
Frame = +3
Query: 113 LNASAWYNLGILHKAEGRV---LEAVECFQAANSLEETAPIEPF 153
LNASAWYNLG+ HKAEG++ +EA ECFQAA+SLEE+ P+EPF
Sbjct: 3 LNASAWYNLGLFHKAEGKISSLVEATECFQAAHSLEESTPLEPF 134
>TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl glucosamine
transferase {Arabidopsis thaliana}, partial (94%)
Length = 3465
Score = 50.1 bits (118), Expect = 4e-07
Identities = 37/138 (26%), Positives = 64/138 (45%), Gaps = 8/138 (5%)
Frame = +2
Query: 12 EIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDA 71
+ W +LA AY+ + +A C ++ A +G + +A+GL +EA + +A
Sbjct: 698 DAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEAYSCYLEA 877
Query: 72 LNIDPRHVPSLISTAVALRRWSNQSDPVVRS--------FLMDALRYDRLNASAWYNLGI 123
L I P + A+A WSN + + S + +A++ A+ NLG
Sbjct: 878 LRIQP-------TFAIA---WSNLAGLFMESGDFNRALQYYKEAVKLKPSFPDAYLNLGN 1027
Query: 124 LHKAEGRVLEAVECFQAA 141
++KA G EA+ C+Q A
Sbjct: 1028VYKALGMPQEAIACYQHA 1081
Score = 38.9 bits (89), Expect = 8e-04
Identities = 27/99 (27%), Positives = 49/99 (49%)
Frame = +2
Query: 51 IGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRY 110
+G +++A G+ +EA+ ++ AL P + + + A ++ Q D + + A+
Sbjct: 1019 LGNVYKALGMPQEAIACYQHALQTRPNYGMAYGNLA-SIHYEQGQLDMAILHY-KQAIAC 1192
Query: 111 DRLNASAWYNLGILHKAEGRVLEAVECFQAANSLEETAP 149
D A+ NLG K GRV EA++C+ SL+ P
Sbjct: 1193 DPRFLEAYNNLGNALKDVGRVEEAIQCYNQCLSLQPNHP 1309
>TC89875 similar to GP|20259245|gb|AAM14358.1 putative N-terminal
acetyltransferase {Arabidopsis thaliana}, partial (38%)
Length = 1170
Score = 42.4 bits (98), Expect = 7e-05
Identities = 29/103 (28%), Positives = 48/103 (46%), Gaps = 2/103 (1%)
Frame = +3
Query: 49 HVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPV-VRSFLMDA 107
HV G ++ + Y+EA+K +R+AL IDP ++ L ++ + + S V R L+
Sbjct: 345 HVFGLLYRSDREYREAIKCYRNALRIDPENIEILRDLSLLQAQMRDLSGFVETRQQLLTL 524
Query: 108 LRYDRLNASAWYNLGILHKAEGRVLEAVECFQA-ANSLEETAP 149
R+N W + H +AVE +A +LE P
Sbjct: 525 KPNHRMN---WIGFSVAHHLNSNASKAVEILEAYEGTLENDHP 644
Score = 32.3 bits (72), Expect = 0.076
Identities = 19/81 (23%), Positives = 38/81 (46%)
Frame = +3
Query: 61 YKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYN 120
YK+ +KA L P H +L + L +S+ + L+ D + W+
Sbjct: 177 YKKGLKAADAILKKFPDHGETLSMKGLTLNCMDRKSEAY--ELVRQGLKNDLKSHVCWHV 350
Query: 121 LGILHKAEGRVLEAVECFQAA 141
G+L++++ EA++C++ A
Sbjct: 351 FGLLYRSDREYREAIKCYRNA 413
>TC79338 homologue to GP|20259245|gb|AAM14358.1 putative N-terminal
acetyltransferase {Arabidopsis thaliana}, partial (24%)
Length = 904
Score = 42.4 bits (98), Expect = 7e-05
Identities = 29/105 (27%), Positives = 50/105 (47%), Gaps = 2/105 (1%)
Frame = +3
Query: 49 HVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPV-VRSFLMDA 107
HV G ++ + Y+EA+K +R+AL IDP ++ L ++ + + S V R L+
Sbjct: 495 HVYGLLYRSDREYREAIKCYRNALRIDPDNIEILRDLSLLQAQMRDLSGFVETRQQLLTL 674
Query: 108 LRYDRLNASAWYNLGILHKAEGRVLEAVECFQA-ANSLEETAPIE 151
R+N W + H +A+E +A +LE+ P E
Sbjct: 675 KSNHRMN---WIGFAVSHHLNSNASKAIEILEAYEGTLEDDYPPE 800
Score = 32.0 bits (71), Expect = 0.100
Identities = 19/81 (23%), Positives = 38/81 (46%)
Frame = +3
Query: 61 YKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYN 120
YK+ +KA L P H +L + L +S+ + L+ D + W+
Sbjct: 327 YKKGLKAADAILKKFPDHGETLSMKGLTLNCMDRKSEAY--ELVRQGLKNDLKSHVCWHV 500
Query: 121 LGILHKAEGRVLEAVECFQAA 141
G+L++++ EA++C++ A
Sbjct: 501 YGLLYRSDREYREAIKCYRNA 563
>TC87394 similar to GP|3582779|gb|AAC69180.1| peroxisomal targeting sequence
1 receptor {Nicotiana tabacum}, partial (65%)
Length = 1656
Score = 33.9 bits (76), Expect = 0.026
Identities = 26/99 (26%), Positives = 46/99 (46%)
Frame = +3
Query: 50 VIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALR 109
V+G ++ Y +A+ AF AL + P+ SL + A + S QS + ++ AL
Sbjct: 789 VLGVLYNLSREYDKAIAAFEQALKLKPQDY-SLWNKLGATQANSVQSADAIAAY-QQALD 962
Query: 110 YDRLNASAWYNLGILHKAEGRVLEAVECFQAANSLEETA 148
AW N+GI + +G E++ + A ++ A
Sbjct: 963 LKPNYVRAWANMGISYANQGMYDESIRYYVRALAMNPKA 1079
>TC92121 weakly similar to GP|10177302|dbj|BAB10563.
gene_id:MDC12.18~pir||F69210~similar to unknown protein
{Arabidopsis thaliana}, partial (76%)
Length = 845
Score = 31.2 bits (69), Expect = 0.17
Identities = 24/128 (18%), Positives = 54/128 (41%)
Frame = +3
Query: 14 WHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALN 73
WH L + ++ ++ L + A + + +G + +A + ++ AL+
Sbjct: 159 WHQLGLHSLCAREFKTSQKYLKAAVACDKGCSYAWSNLGVSLQLSEEQSQAEEVYKWALS 338
Query: 74 IDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLE 133
+ + I + + + + + ++ +L A A+ NLG++ AEG + E
Sbjct: 339 LATKQEAHAILSNMGILYRQQKKYELAKAMFTKSLELQPGYAPAFNNLGLVFIAEGLLEE 518
Query: 134 AVECFQAA 141
A CF+ A
Sbjct: 519 AKHCFEKA 542
>TC89957 weakly similar to PIR|D96810|D96810 hypothetical protein T11I11.6
[imported] - Arabidopsis thaliana, partial (51%)
Length = 1843
Score = 29.6 bits (65), Expect = 0.50
Identities = 28/99 (28%), Positives = 48/99 (48%), Gaps = 3/99 (3%)
Frame = +3
Query: 37 SKAFRQYTASRCHVIGTM-HEAKGLYKEAVKAFRDALNIDP--RHVPSLISTAVALRRWS 93
+K F T++ +I + + A G ++EAVK + A +DP R V +++ A A+
Sbjct: 1140 NKIFGMATSAYLSMISALVYLASGRFEEAVKTSQQADRVDPSNREVNAVLRRAKAV---- 1307
Query: 94 NQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVL 132
S + + L A ++ + A A YN G+ H VL
Sbjct: 1308 -TSSRMSGNLLFKASKF--MEACAVYNEGLDHDPHNSVL 1415
>AW586685 similar to PIR|T01081|T01 hypothetical protein T10P11.3.2 -
Arabidopsis thaliana, partial (22%)
Length = 651
Score = 28.1 bits (61), Expect = 1.4
Identities = 16/45 (35%), Positives = 24/45 (52%)
Frame = +2
Query: 34 LSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRH 78
LS++ AF+ H+ HE KG A++ R AL++DP H
Sbjct: 446 LSRAIAFKA-DLHLLHLRAAFHEHKGDVLSALRDCRAALSVDPNH 577
>TC79242
Length = 1802
Score = 26.2 bits (56), Expect = 5.5
Identities = 12/36 (33%), Positives = 20/36 (55%)
Frame = +3
Query: 112 RLNASAWYNLGILHKAEGRVLEAVECFQAANSLEET 147
RLN +W +L +LH+ R+ + F A+S +T
Sbjct: 957 RLNCVSWLSLRLLHRIGERLCFTSDLFDNADSATKT 1064
>AL381331 weakly similar to GP|14209525|dbj contains ESTs C73715(E20247)
C99497(E20247)~similar to Arabidopsis thaliana
chromosome 2 At2g20670~, partial (14%)
Length = 475
Score = 25.8 bits (55), Expect = 7.2
Identities = 20/63 (31%), Positives = 26/63 (40%)
Frame = -2
Query: 34 LSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWS 93
+S SK FRQ T H H ++ + +F L P L+ VALR WS
Sbjct: 345 ISVSKHFRQLTTGCRH----KHSISDMFISLIASFVACLTFVS---PELVLHKVALRSWS 187
Query: 94 NQS 96
S
Sbjct: 186 CSS 178
>TC87659 similar to GP|10177021|dbj|BAB10259. contains similarity to sorting
nexin~gene_id:MQJ2.4 {Arabidopsis thaliana}, partial
(70%)
Length = 1803
Score = 25.4 bits (54), Expect = 9.3
Identities = 13/44 (29%), Positives = 19/44 (42%)
Frame = +1
Query: 30 AEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALN 73
A + S+ FR+ + + T+HE GL AF D N
Sbjct: 1372 ATAAVKASRLFRELNSQTVKHLDTLHEYLGLMLAVHSAFTDRTN 1503
>TC84816 similar to GP|2313789|gb|AAD07729.1| H. pylori predicted coding
region HP0659 {Helicobacter pylori 26695}, partial (3%)
Length = 807
Score = 25.4 bits (54), Expect = 9.3
Identities = 11/32 (34%), Positives = 15/32 (46%)
Frame = +2
Query: 19 YAYISLSQWHDAEVCLSKSKAFRQYTASRCHV 50
Y + SLS + + SK R YT CH+
Sbjct: 425 YNFDSLSNNDQQNLLVRSSKILRTYTEGNCHI 520
>BQ144111
Length = 748
Score = 25.4 bits (54), Expect = 9.3
Identities = 8/17 (47%), Positives = 12/17 (70%)
Frame = -2
Query: 46 SRCHVIGTMHEAKGLYK 62
SRCH++G +H K Y+
Sbjct: 414 SRCHIVGHIHRNKSHYR 364
>AW776563 similar to GP|21304447|em HOBBIT protein {Arabidopsis thaliana},
partial (15%)
Length = 342
Score = 25.4 bits (54), Expect = 9.3
Identities = 19/68 (27%), Positives = 36/68 (52%), Gaps = 3/68 (4%)
Frame = +3
Query: 61 YKEAVKAFRDALNIDPRHVPSLISTA---VALRRWSNQSDPVVRSFLMDALRYDRLNASA 117
++ A+K F+ A+ ++PR + VAL + N ++ + ALR D + +A
Sbjct: 9 HETALKNFQRAVQLNPRFAYAHTLCGHEYVAL*DFENG----IKCY-QSALRVDERHYNA 173
Query: 118 WYNLGILH 125
WY LG+++
Sbjct: 174 WYGLGMVY 197
>TC91796 GP|23498903|emb|CAD50981. hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 836
Score = 25.4 bits (54), Expect = 9.3
Identities = 9/28 (32%), Positives = 15/28 (53%)
Frame = -3
Query: 13 IWHDLAYAYISLSQWHDAEVCLSKSKAF 40
+W + +A S +HD +C+S K F
Sbjct: 378 VWINSLFAAYITSSYHDKSLCISSHKLF 295
>AW775312 similar to GP|20466732|gb unknown protein {Arabidopsis thaliana},
partial (46%)
Length = 655
Score = 25.4 bits (54), Expect = 9.3
Identities = 12/36 (33%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Frame = +2
Query: 13 IWHDLAY-AYISLSQWHDAEVCLSKSKAFRQYTASR 47
+WH L +Y SL QW + C+S + F +S+
Sbjct: 197 LWHRLPTGSYRSLHQWSNLRRCISSTFGFSSIYSSK 304
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,660,601
Number of Sequences: 36976
Number of extensions: 54021
Number of successful extensions: 276
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 266
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 271
length of query: 153
length of database: 9,014,727
effective HSP length: 88
effective length of query: 65
effective length of database: 5,760,839
effective search space: 374454535
effective search space used: 374454535
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0082b.2


