

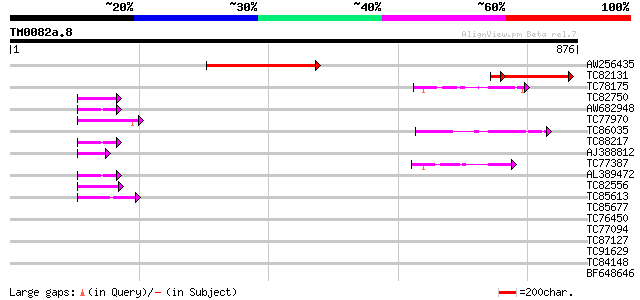
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082a.8
(876 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW256435 weakly similar to PIR|T06657|T066 hypothetical protein ... 238 1e-62
TC82131 similar to PIR|T06657|T06657 hypothetical protein T6G15.... 208 5e-62
TC78175 similar to GP|1527001|gb|AAB07724.1| Ipomoea nil Pn47p, ... 57 3e-08
TC82750 similar to GP|21592759|gb|AAM64708.1 unknown {Arabidopsi... 50 3e-06
AW682948 weakly similar to PIR|E96525|E96 protein T1N15.21 [impo... 50 5e-06
TC77970 similar to GP|20197686|gb|AAM15203.1 expressed protein {... 49 6e-06
TC86035 similar to PIR|E71435|E71435 probable triacylglycerol li... 47 2e-05
TC88217 weakly similar to GP|18377801|gb|AAL67050.1 unknown prot... 47 4e-05
AJ388812 weakly similar to GP|15810203|g AT5g50170/K6A12_3 {Arab... 46 7e-05
TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19 ... 45 1e-04
AL389472 similar to GP|6648206|gb|A putative GTPase activating p... 44 2e-04
TC82556 similar to GP|21536965|gb|AAM61306.1 putative zinc finge... 43 6e-04
TC85613 weakly similar to EGAD|2627|2550 protein kinase C beta ... 42 8e-04
TC85677 weakly similar to GP|6457331|gb|AAF09479.1| phytoalexin-... 40 0.004
TC76450 similar to GP|17065130|gb|AAL32719.1 putative protein {A... 40 0.004
TC77094 weakly similar to PIR|A96608|A96608 hypothetical protein... 40 0.005
TC87127 similar to GP|12324745|gb|AAG52327.1 unknown protein; 38... 39 0.007
TC91629 similar to PIR|A96608|A96608 hypothetical protein F25P12... 39 0.007
TC84148 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unk... 37 0.043
BF648646 similar to GP|20161014|db hypothetical protein~similar ... 35 0.12
>AW256435 weakly similar to PIR|T06657|T066 hypothetical protein T6G15.100 -
Arabidopsis thaliana, partial (6%)
Length = 563
Score = 238 bits (606), Expect = 1e-62
Identities = 121/176 (68%), Positives = 146/176 (82%)
Frame = +3
Query: 305 NEACSEQRNTEEFHSHESDTENGHTSEPSTQTSGEELSNQLFWRDFTNVINVNVVQKLGL 364
NEA SEQRN EEF S +S+TENGH E S Q S EELSNQ FW++F+ V+N NVVQKLGL
Sbjct: 6 NEAPSEQRNMEEFRSCDSETENGHALEASPQASEEELSNQRFWKNFSKVVNANVVQKLGL 185
Query: 365 TVPGKFKWDGLEFLDKIGSQSQNIAEATYIQSGLAMPRGIDDTEDKTSGKPAIAAIQSAL 424
+VP K KWDGLEFL+KIGSQSQ++AE YI+SGLA+P G D + +KT G+P+IAAIQS+L
Sbjct: 186 SVPEKLKWDGLEFLNKIGSQSQDVAEDIYIESGLAIPEGTDISYNKTCGQPSIAAIQSSL 365
Query: 425 PEVKKATESLMRQTDSILGGLMLLAATVSKMKDEGSSSEERKIEEDSAKVGGNDIQ 480
PEVKKATE+LM+QT+SILGGLMLL AT S+MKDE SS+ERK +EDS + G+DIQ
Sbjct: 366 PEVKKATETLMKQTESILGGLMLLTATDSRMKDERRSSDERKTKEDSTE*VGSDIQ 533
>TC82131 similar to PIR|T06657|T06657 hypothetical protein T6G15.100 -
Arabidopsis thaliana, partial (14%)
Length = 890
Score = 208 bits (530), Expect(2) = 5e-62
Identities = 98/111 (88%), Positives = 107/111 (96%)
Frame = +1
Query: 761 KIKDSWRVVNHRDIIPTVPRLMGYCHVNQPVFLAAGVLRNSLENKDILGDGYEGDVLGES 820
K+K +WR+VNHRDIIPT+PRLMGYCHVNQP+FLAAGV NSLENKDILGDGYEGDVLGES
Sbjct: 61 KLKIAWRIVNHRDIIPTIPRLMGYCHVNQPLFLAAGVSTNSLENKDILGDGYEGDVLGES 240
Query: 821 TPDVIVSEFMKGEKELIEKLLQTEINIFRSIRDGTAFMQHMEDFYYVTLLE 871
TPDVIV+EFMKGE ELIEKLLQTEINIFRSIRDG+A+MQHMEDFYY+TLLE
Sbjct: 241 TPDVIVNEFMKGEMELIEKLLQTEINIFRSIRDGSAYMQHMEDFYYITLLE 393
Score = 48.9 bits (115), Expect(2) = 5e-62
Identities = 21/23 (91%), Positives = 23/23 (99%)
Frame = +3
Query: 743 NFGSPRVGNKRFAEVYNEKIKDS 765
+FGSPRVGNKRFAEVYNEK+KDS
Sbjct: 6 SFGSPRVGNKRFAEVYNEKVKDS 74
>TC78175 similar to GP|1527001|gb|AAB07724.1| Ipomoea nil Pn47p, partial
(32%)
Length = 1153
Score = 57.0 bits (136), Expect = 3e-08
Identities = 54/201 (26%), Positives = 92/201 (44%), Gaps = 21/201 (10%)
Frame = +1
Query: 624 KQEVQVHSGFLSAY-----------DSVRTRIISLI-RLAIGYVDDHFEPLHKWHIYVTG 671
K +VQ+H+GF S Y S R ++++ I RL Y ++ I VTG
Sbjct: 586 KSDVQLHNGFYSLYTSDNSSLPLADSSARKQVLNEISRLVELYKNEEIS------ITVTG 747
Query: 672 HSLGGALATLLALELSSNQLTNMVDYVEEPKTHHIRQIMIANYGRISAMALSCPIFDNNA 731
HSLGGALAT+ ++++ +N+ + + KT CP
Sbjct: 748 HSLGGALATISSMDIVANKFN--IPKGQPQKT--------------------CP------ 843
Query: 732 HLRGAISITMYNFGSPRVGNKRFAEVYNEKIK-DSWRVVNHRDIIPTVPRLMGYCHVNQP 790
+T++ FGSPRVGN F +++++ + + N+ DI+P+ RL Y V +
Sbjct: 844 -------VTLFAFGSPRVGNSNFEKIFSDNNDLRALFIRNNNDIVPSSLRL-AYSKVGEE 999
Query: 791 V--------FLAAGVLRNSLE 803
+ +L +GV +++E
Sbjct: 1000LEIDTEKSKYLKSGVSEHNME 1062
>TC82750 similar to GP|21592759|gb|AAM64708.1 unknown {Arabidopsis
thaliana}, partial (95%)
Length = 623
Score = 50.4 bits (119), Expect = 3e-06
Identities = 25/68 (36%), Positives = 39/68 (56%)
Frame = +3
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
TSDPYVV ++ +QT K+ + P WNEE F + P L + +D +L+ +M
Sbjct: 135 TSDPYVVLKLGNQTAKTKVINSCLNPVWNEELNFTL-TEPLGVLNLEVFDKDLLKADDKM 311
Query: 166 GNAGVDLE 173
GNA ++L+
Sbjct: 312 GNAFINLQ 335
>AW682948 weakly similar to PIR|E96525|E96 protein T1N15.21 [imported] -
Arabidopsis thaliana, partial (72%)
Length = 576
Score = 49.7 bits (117), Expect = 5e-06
Identities = 25/68 (36%), Positives = 37/68 (53%)
Frame = +3
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
TSDPYV+ M+ QT+K+ + P WNEE T IK N P+ + D + +M
Sbjct: 78 TSDPYVLVTMEEQTLKTAVVNDNCHPEWNEELTLYIK-DVNTPIHLIVCDKDTFTVDDKM 254
Query: 166 GNAGVDLE 173
G A +D++
Sbjct: 255 GEADIDIK 278
>TC77970 similar to GP|20197686|gb|AAM15203.1 expressed protein {Arabidopsis
thaliana}, partial (97%)
Length = 2062
Score = 49.3 bits (116), Expect = 6e-06
Identities = 31/115 (26%), Positives = 52/115 (44%), Gaps = 12/115 (10%)
Frame = +2
Query: 105 GTSDPYVVFQMDSQTV---KSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAP 161
G SDPYV ++ + K+ +K P WNEEF +K P + LQ+ +D V
Sbjct: 1007 GASDPYVKLKLTDDKMPSKKTTVKHKNLNPEWNEEFNLVVKDPETQVLQLNVYDWEQVGK 1186
Query: 162 HKRMGNAGVDLEWLCDGDVHEILVEL---------EGMGGGGKVQLEVKYKTFDE 207
H +MG + L+ + + ++L + G++ +EV YK +E
Sbjct: 1187 HDKMGMNVITLKEVSPEEPKRFTLDLLKTMDPNDAQNEKSRGQIVVEVTYKPLNE 1351
>TC86035 similar to PIR|E71435|E71435 probable triacylglycerol lipase -
Arabidopsis thaliana, partial (57%)
Length = 1783
Score = 47.4 bits (111), Expect = 2e-05
Identities = 56/215 (26%), Positives = 89/215 (41%), Gaps = 6/215 (2%)
Frame = +3
Query: 628 QVHSGFLSAYDSVRTRIISLIRLAIGYVDDHFEPLHKWH---IYVTGHSLGGALATLLAL 684
+V GF+S Y + + SL + V E L+K I VTGHSLG LA L+A
Sbjct: 1005 KVECGFMSLYKTKGAHVQSLSESVVEEVRRLIE-LYKGEELSITVTGHSLGATLALLVAE 1181
Query: 685 ELSSNQLTNMVDYVEEPKTHHIRQIMIANYGRISAMALSCPIFDNNAHLRGAISITMYNF 744
E IS A + P + +++F
Sbjct: 1182 E-------------------------------ISTCAPNVP------------PVAVFSF 1232
Query: 745 GSPRVGNKRFAEVYNEKIKDSWRVVNHRDIIPTVPRLMGYCHVNQPV---FLAAGVLRNS 801
G PRVGN+ F E +K R+VN +D+I VP + + + + + +GV+ +
Sbjct: 1233 GGPRVGNRAFGEHLEKKNVKVLRIVNTQDVITRVPGIFLSEELEEKIKNSKVVSGVV-DM 1409
Query: 802 LENKDILGDGYEGDVLGESTPDVIVSEFMKGEKEL 836
LE LG + G L +T +S ++K + ++
Sbjct: 1410 LEENTPLGYSHVGTELRVNTK---MSPYLKPDADI 1505
>TC88217 weakly similar to GP|18377801|gb|AAL67050.1 unknown protein
{Arabidopsis thaliana}, partial (94%)
Length = 857
Score = 46.6 bits (109), Expect = 4e-05
Identities = 22/67 (32%), Positives = 38/67 (55%)
Frame = +3
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDPYVV +M Q +K+ + P WNE+ T +I P+ P+ + +D + + +M
Sbjct: 153 SSDPYVVIKMAKQKLKTRVVKKNLNPEWNEDLTLSIS-DPHTPIHLYVYDKDTFSLDDKM 329
Query: 166 GNAGVDL 172
G+A D+
Sbjct: 330 GDAEFDI 350
>AJ388812 weakly similar to GP|15810203|g AT5g50170/K6A12_3 {Arabidopsis
thaliana}, partial (14%)
Length = 673
Score = 45.8 bits (107), Expect = 7e-05
Identities = 22/51 (43%), Positives = 29/51 (56%)
Frame = +2
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWD 155
G SDPYVVF + QT S++K T +P WNE F+ P L V ++D
Sbjct: 338 GLSDPYVVFTCNGQTRSSSVKLETSDPQWNEILEFDAMEEPPSVLXVESFD 490
>TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19
{Arabidopsis thaliana}, partial (75%)
Length = 1793
Score = 45.1 bits (105), Expect = 1e-04
Identities = 46/174 (26%), Positives = 73/174 (41%), Gaps = 13/174 (7%)
Frame = +3
Query: 622 DFKQE--VQVHSGFLSAYD-----------SVRTRIISLIRLAIGYVDDHFEPLHKWHIY 668
+FK + ++V +GF Y S R +++S I+ + + K I
Sbjct: 864 NFKNDPSIKVETGFYDLYTKKEQSCTYCSFSAREQVLSEIKRLLQFYQGE-----KISIT 1028
Query: 669 VTGHSLGGALATLLALELSSNQLTNMVDYVEEPKTHHIRQIMIANYGRISAMALSCPIFD 728
VTGHSLG ALA L A +++ + N+++ + KT ++
Sbjct: 1029VTGHSLGAALAVLSAYDIAELGV-NIIE--DGDKTTNV---------------------- 1133
Query: 729 NNAHLRGAISITMYNFGSPRVGNKRFAEVYNEKIKDSWRVVNHRDIIPTVPRLM 782
IT+Y+F PRVGN F E E R+ N D +PTVP ++
Sbjct: 1134---------PITVYSFAGPRVGNLHFKERCEELGVKVLRIHNIHDKVPTVPGII 1268
>AL389472 similar to GP|6648206|gb|A putative GTPase activating protein
{Arabidopsis thaliana}, partial (24%)
Length = 487
Score = 44.3 bits (103), Expect = 2e-04
Identities = 23/69 (33%), Positives = 36/69 (51%), Gaps = 1/69 (1%)
Frame = +1
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPN-KPLQVAAWDANLVAPHKR 164
TSDPYV+ + Q+VK+ + P WNE +I P N PL++ +D +
Sbjct: 286 TSDPYVILSLGHQSVKTRVIKNNLNPVWNESLMLSI--PENIPPLKIIVYDKDSFKNDDF 459
Query: 165 MGNAGVDLE 173
MG A +D++
Sbjct: 460 MGEAEIDIQ 486
>TC82556 similar to GP|21536965|gb|AAM61306.1 putative zinc finger and C2
domain protein {Arabidopsis thaliana}, partial (40%)
Length = 756
Score = 42.7 bits (99), Expect = 6e-04
Identities = 22/70 (31%), Positives = 39/70 (55%)
Frame = +3
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDPYVV + +QTV++++ P WNEE ++ + L++ +D + + M
Sbjct: 39 SSDPYVVLNLGTQTVQTSVMRSNLNPVWNEEHMLSVPEHYGQ-LKLKVFDHDTFSADDIM 215
Query: 166 GNAGVDLEWL 175
G A +DL+ L
Sbjct: 216 GEADIDLQSL 245
>TC85613 weakly similar to EGAD|2627|2550 protein kinase C beta {Homo
sapiens}, partial (3%)
Length = 584
Score = 42.4 bits (98), Expect = 8e-04
Identities = 33/102 (32%), Positives = 49/102 (47%), Gaps = 4/102 (3%)
Frame = +1
Query: 105 GTSDPYVVFQMDSQ-TVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHK 163
G SDPYV + ++ K GT P WNE FTFN+ + L+V D V
Sbjct: 142 GKSDPYVELWIKKDYKQRTTTKKGTVNPEWNETFTFNVDGDHHLYLKVLDDD---VVSDD 312
Query: 164 RMGNAGVDLEWLCDGDVHEILVELE---GMGGGGKVQLEVKY 202
++G A VDL+ + D V+L G+ G +QL +++
Sbjct: 313 KIGEAKVDLKQVYDSHYLSTDVKLPALLGLTNHGFIQLVLEF 438
>TC85677 weakly similar to GP|6457331|gb|AAF09479.1| phytoalexin-deficient 4
protein {Arabidopsis thaliana}, partial (21%)
Length = 2227
Score = 40.0 bits (92), Expect = 0.004
Identities = 45/188 (23%), Positives = 76/188 (39%), Gaps = 9/188 (4%)
Frame = +3
Query: 598 LDPKGVFVGVRYPIARLNPERIGGDFKQEVQVHSGFL----SAYDSVRTRIISLIRLAIG 653
L+P GV R N E ++ V+VHSG L S ++S++ +++ ++
Sbjct: 351 LEPLESIGGVPLFSTRRNKEE-----EEPVKVHSGMLNLFSSLFNSIQNQVLGIL----- 500
Query: 654 YVDDHFEPLHKWHIYVTGHSLGGALATLLALELSSNQLTNMVDYVEEPKTHHIRQIMIAN 713
E + +TGHS+GGA+A+L L L S
Sbjct: 501 ------ENTDAKSLVITGHSIGGAIASLCTLWLLS------------------------- 587
Query: 714 YGRISAMALSCPIFDNNAHLRGAISITMYNFGSPRVGNKRFAEVYNEKIKDSW-----RV 768
I++++ S P+ FGSP +GNK F++ + ++ W V
Sbjct: 588 --YINSISSSLPVM-------------CITFGSPLLGNKSFSQAIS---REKWGGNFCHV 713
Query: 769 VNHRDIIP 776
V+ DI+P
Sbjct: 714 VSKHDIMP 737
>TC76450 similar to GP|17065130|gb|AAL32719.1 putative protein {Arabidopsis
thaliana}, partial (83%)
Length = 2171
Score = 40.0 bits (92), Expect = 0.004
Identities = 18/52 (34%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Frame = +1
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNI-KLPPNKPLQVAAWD 155
GTSDPY + ++ S++ G++ P W EEF F++ +LP + + WD
Sbjct: 535 GTSDPYTIITCGNEKRFSSMVPGSRNPMWGEEFNFSVDELPVQINVTIYDWD 690
>TC77094 weakly similar to PIR|A96608|A96608 hypothetical protein F25P12.93
[imported] - Arabidopsis thaliana, partial (57%)
Length = 1745
Score = 39.7 bits (91), Expect = 0.005
Identities = 71/312 (22%), Positives = 112/312 (35%), Gaps = 11/312 (3%)
Frame = +1
Query: 479 IQCLPKIPSSENGSVLDDKKAEEMRELFSTAETAMEAWAMLATSLGHASFIKSEFEKICF 538
+ C+ + S N +L K E+ + S A A +A L + + E + F
Sbjct: 430 LSCIGNLDSRVNMDIL---KREDNKYYVSLAMMASKAVYENEAFLKYTIKYDWKMEYVGF 600
Query: 539 LD-----NAPTDTQVAIWRDSMRRR--LVVAFRGTEQSQWKDLITDLMIVPAGLASSSFS 591
D TQV I D + R VVAFRGTE TDL I G+
Sbjct: 601 FDCWNEYQERATTQVLILLDKFKDRDTYVVAFRGTEPFDADAWCTDLDISWYGIPG--VG 774
Query: 592 SAQASILDPKGVFVGVRYPIARLNPERIGGDFKQEVQVHSGFLSAYDSVRTRIISLIRLA 651
A + G+ + +P E++ AY +R ++R
Sbjct: 775 RAHGGFMKALGLQKNLGWPKEIERDEKLAP-------------LAYYVIR----DILRKG 903
Query: 652 IGYVDDHFEPLHKWHIYVTGHSLGGALATLLALELSSNQLTNMVDYVEEPKTHHIRQIMI 711
+ P K+ +TGHSLGGALA L + + +++ +E
Sbjct: 904 LSE-----NPNAKF--IITGHSLGGALAILFPTIMFLHDEKLLIERLEG----------- 1029
Query: 712 ANYGRISAMALSCPIFDNNAHLRGAISITMYNFGSPRVGNKRFAEVYNEKIKDS----WR 767
+Y FG PRVG++R+ + +K+K++ R
Sbjct: 1030-----------------------------IYTFGQPRVGDERYTQYMTQKMKENRITYCR 1122
Query: 768 VVNHRDIIPTVP 779
V DI+P +P
Sbjct: 1123FVYCNDIVPRLP 1158
>TC87127 similar to GP|12324745|gb|AAG52327.1 unknown protein; 3866-2463
{Arabidopsis thaliana}, partial (76%)
Length = 747
Score = 39.3 bits (90), Expect = 0.007
Identities = 22/69 (31%), Positives = 37/69 (52%), Gaps = 1/69 (1%)
Frame = +3
Query: 106 TSDPYVVFQMD-SQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
+SDPYVV + Q +K+ + P WNEE T +I+ P+ + +D + +
Sbjct: 99 SSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIR-DVRVPICLTVFDKDTFFVDDK 275
Query: 165 MGNAGVDLE 173
MG+A +DL+
Sbjct: 276 MGDAEIDLK 302
>TC91629 similar to PIR|A96608|A96608 hypothetical protein F25P12.93
[imported] - Arabidopsis thaliana, partial (67%)
Length = 1291
Score = 39.3 bits (90), Expect = 0.007
Identities = 30/115 (26%), Positives = 45/115 (39%), Gaps = 4/115 (3%)
Frame = +3
Query: 669 VTGHSLGGALATLLALELSSNQLTNMVDYVEEPKTHHIRQIMIANYGRISAMALSCPIFD 728
+TGHSLGG LA L L ++ +M+D ++
Sbjct: 558 LTGHSLGGTLAILFVAMLIFHEEEDMLDKLQG---------------------------- 653
Query: 729 NNAHLRGAISITMYNFGSPRVGNKRFAEVYNEKIK----DSWRVVNHRDIIPTVP 779
+Y FG PRVG+++F E K+K +R V D++P VP
Sbjct: 654 ------------VYTFGQPRVGDEKFGEFMKSKLKKYDVKYFRYVYSNDMVPRVP 782
>TC84148 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unknown
protein {Arabidopsis thaliana}, partial (16%)
Length = 840
Score = 36.6 bits (83), Expect = 0.043
Identities = 28/133 (21%), Positives = 55/133 (41%)
Frame = +1
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
GTSDP+V + ++ + T P W++ F L PL + D N + P
Sbjct: 169 GTSDPFVRVNYGNLKKRTKVVHKTINPRWDQTLEF---LDDGSPLTLHVKDHNALLPTSS 339
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGGGGKVQLEVKYKTFDEIDDEKKWWKMPFVSDFL 224
+G V+ + L + + L+G+ G E+ + ++ + + F
Sbjct: 340 IGECVVEYQSLPPNQTSDKWIPLQGVKSG-----EIHIQIARKVPEIQTRQSPDFEPSLT 504
Query: 225 KINGFDSALKKVT 237
K++ S +K++T
Sbjct: 505 KLHQSPSQIKEMT 543
>BF648646 similar to GP|20161014|db hypothetical protein~similar to
PK1-related protein kinase 2 {Oryza sativa (japonica
cultivar-group), partial (24%)
Length = 655
Score = 35.0 bits (79), Expect = 0.12
Identities = 42/156 (26%), Positives = 67/156 (42%), Gaps = 13/156 (8%)
Frame = +1
Query: 559 LVVAFRGTEQSQWKDLITDLMIVPAGLASSSFSSAQ--ASILDPKGVFVGVRYPIARLNP 616
+V+A RGTE + DLITD GL SA+ A +++ + + +A P
Sbjct: 172 VVIAIRGTETPE--DLITD------GLCKECTLSAEDLAGLINCNHIHSDIHKNVASSFP 327
Query: 617 ERIGGDFKQEVQVHSGFLSAYDSVRTRI-----------ISLIRLAIGYVDDHFEPLHKW 665
HSG + A + +I L+ +G+ + F +
Sbjct: 328 H----------YGHSGIVEAARELYMQIEGNPGEHDTESYGLLSKLLGFGCECFG----Y 465
Query: 666 HIYVTGHSLGGALATLLALELSSNQLTNMVDYVEEP 701
++ + GHSLGGA+A LL L+L N+ N+ Y P
Sbjct: 466 NVRIVGHSLGGAIAALLGLQL-YNRYPNLHVYSYGP 570
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,837,256
Number of Sequences: 36976
Number of extensions: 326305
Number of successful extensions: 1850
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 1818
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1849
length of query: 876
length of database: 9,014,727
effective HSP length: 105
effective length of query: 771
effective length of database: 5,132,247
effective search space: 3956962437
effective search space used: 3956962437
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0082a.8


