

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
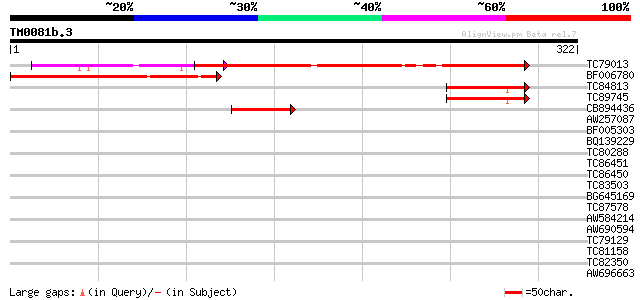
Query= TM0081b.3
(322 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79013 similar to GP|21537154|gb|AAM61495.1 unknown {Arabidopsi... 176 1e-44
BF006780 similar to GP|10177469|dbj gene_id:MQB2.23~pir||T06676~... 154 3e-38
TC84813 similar to PIR|T47503|T47503 hypothetical protein F9K21.... 60 1e-09
TC89745 similar to PIR|T47503|T47503 hypothetical protein F9K21.... 60 1e-09
CB894436 similar to GP|10177469|dbj gene_id:MQB2.23~pir||T06676~... 43 2e-04
AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapi... 40 0.002
BF005303 weakly similar to GP|20197705|gb| putative RING-H2 zinc... 39 0.002
BQ139229 weakly similar to GP|20453199|gb AT3g18290/MIE15_8 {Ara... 36 0.023
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 35 0.030
TC86451 similar to GP|21537318|gb|AAM61659.1 unknown {Arabidopsi... 35 0.030
TC86450 similar to GP|21537318|gb|AAM61659.1 unknown {Arabidopsi... 35 0.030
TC83503 weakly similar to PIR|T51851|T51851 RING-H2 finger prote... 35 0.039
BG645169 similar to PIR|D86157|D86 hypothetical protein AAF02892... 34 0.066
TC87578 similar to PIR|T12113|T12113 transcription factor - fava... 34 0.087
AW584214 similar to GP|7363283|db Similar to Arabidopsis thalian... 34 0.087
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 33 0.11
TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.... 33 0.11
TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidops... 33 0.15
TC82350 similar to GP|9837385|gb|AAG00554.1| retinitis pigmentos... 33 0.19
AW696663 weakly similar to GP|17827193|dbj ORF1 {TT virus}, part... 32 0.33
>TC79013 similar to GP|21537154|gb|AAM61495.1 unknown {Arabidopsis
thaliana}, partial (42%)
Length = 1471
Score = 176 bits (446), Expect = 1e-44
Identities = 94/191 (49%), Positives = 118/191 (61%), Gaps = 1/191 (0%)
Frame = +3
Query: 106 SSGKDFSGSSSNSISTSCSGNDSEEEDDDGCLDDWEAVADALYANDKSHSVVSESPA-EH 164
SSG FSG+ + + EEE DDGCLDDWEAVADAL A++K + + P E
Sbjct: 558 SSGGCFSGNITEEEEDDEEEVEQEEEVDDGCLDDWEAVADALAADEKHRASPQDEPVVET 737
Query: 165 EAECRYTVLEDDKNPRVDFSKADFKSEVPESKPNRRAWKPDDTLRPRCLPNLSKQRNSPL 224
++ C T D + SK VP + N +AW+ DD RP+ LPNLSKQ + P
Sbjct: 738 DSSCEVT---DGLSSGCLDSKPGSGGTVPRASGNGKAWRADDAFRPQTLPNLSKQHSMP- 905
Query: 225 NSNWRGSRKTVPWAWQTIISQPSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEAD 284
N + G VPW+ ++ PS CPIC EDLD+TD+SFLPC+CGF LCLFCHK+ILE D
Sbjct: 906 NPRFVGG---VPWSCMSV---PSNCPICCEDLDLTDTSFLPCNCGFRLCLFCHKRILEQD 1067
Query: 285 ARCPSCRKLYD 295
ARCP CRK Y+
Sbjct: 1068ARCPGCRKQYE 1100
Score = 76.3 bits (186), Expect = 2e-14
Identities = 51/119 (42%), Positives = 68/119 (56%), Gaps = 7/119 (5%)
Frame = +2
Query: 13 KKKRTNGSAKLKQIKLDVRREQWLSR---VKKGC--NVDSNGRMDNCPSSKHTASEENRS 67
KKKRTN SAKLKQ K+D RREQWLS+ KKGC +D + + + P K + E +
Sbjct: 206 KKKRTNRSAKLKQYKIDARREQWLSQGAVKKKGCKDGLDDDVHVSSSPVGKQSKREMEKV 385
Query: 68 SCKEMTRKGEDIEGTCIQDSDSRSTINSP--DRSTYSHDESSGKDFSGSSSNSISTSCS 124
+ R+G + + QDSDS S NSP S+ SG +F+GSSS + S+S S
Sbjct: 386 GTR---RRGGEDDVLIHQDSDSESPSNSPISPNSSVLCGNDSGTNFTGSSSGASSSSSS 553
>BF006780 similar to GP|10177469|dbj gene_id:MQB2.23~pir||T06676~similar to
unknown protein {Arabidopsis thaliana}, partial (12%)
Length = 431
Score = 154 bits (390), Expect = 3e-38
Identities = 83/121 (68%), Positives = 98/121 (80%), Gaps = 1/121 (0%)
Frame = +2
Query: 1 MASGFISDPPLS-KKKRTNGSAKLKQIKLDVRREQWLSRVKKGCNVDSNGRMDNCPSSKH 59
MA G ISDPP S KKKRTNGSAKLKQIKLDVRREQWLSRVKKGC+VDS GR+D+ PS+KH
Sbjct: 71 MAPGSISDPPFSSKKKRTNGSAKLKQIKLDVRREQWLSRVKKGCDVDSTGRVDSYPSAKH 250
Query: 60 TASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSSSNSI 119
ASEEN+ S KE+ RKG + +GTCIQ +DS S IN+ +S+Y+HDES F+GS S+S
Sbjct: 251 IASEENQPSYKEIRRKGRE-DGTCIQGNDSGSIINNSIQSSYNHDESR-NGFNGSRSSST 424
Query: 120 S 120
S
Sbjct: 425 S 427
>TC84813 similar to PIR|T47503|T47503 hypothetical protein F9K21.210 -
Arabidopsis thaliana, partial (15%)
Length = 682
Score = 60.1 bits (144), Expect = 1e-09
Identities = 22/53 (41%), Positives = 33/53 (61%), Gaps = 6/53 (11%)
Frame = +2
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL------EADARCPSCRKLYD 295
CP+C E++D+TD PC CG+ +C++C I+ + D RCP+CR YD
Sbjct: 239 CPLCAEEMDLTDQQLKPCRCGYEICVWCWHHIMDMAEKDDTDGRCPACRSPYD 397
>TC89745 similar to PIR|T47503|T47503 hypothetical protein F9K21.210 -
Arabidopsis thaliana, partial (16%)
Length = 605
Score = 60.1 bits (144), Expect = 1e-09
Identities = 22/53 (41%), Positives = 33/53 (61%), Gaps = 6/53 (11%)
Frame = +1
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL------EADARCPSCRKLYD 295
CP+C E++D+TD PC CG+ +C++C I+ E + RCP+CR YD
Sbjct: 148 CPLCAEEMDLTDQQLRPCKCGYQICVWCWHHIMDMAEKDETEGRCPACRSPYD 306
>CB894436 similar to GP|10177469|dbj gene_id:MQB2.23~pir||T06676~similar to
unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 223
Score = 42.7 bits (99), Expect = 2e-04
Identities = 20/36 (55%), Positives = 24/36 (66%)
Frame = +3
Query: 127 DSEEEDDDGCLDDWEAVADALYANDKSHSVVSESPA 162
+ EEE DDGC DD EAVA+AL A +K + E PA
Sbjct: 48 EQEEEGDDGCWDDGEAVAEALAAEEKHRASPQEEPA 155
>AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapiens},
partial (2%)
Length = 466
Score = 39.7 bits (91), Expect = 0.002
Identities = 30/90 (33%), Positives = 46/90 (50%)
Frame = +1
Query: 45 VDSNGRMDNCPSSKHTASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHD 104
VDS+ D+ SS +++ + SS K+ ++SDS S D S+ S
Sbjct: 154 VDSDSDSDS-DSSSSSSTSSSVSSVKKPAVSAAKATPAKKEESDSDS---DSDSSSSSSS 321
Query: 105 ESSGKDFSGSSSNSISTSCSGNDSEEEDDD 134
SS S SSS+S S+S S +D EEE+++
Sbjct: 322 SSSSSSSSSSSSSSSSSSSSNSDDEEEEEE 411
>BF005303 weakly similar to GP|20197705|gb| putative RING-H2 zinc finger
protein {Arabidopsis thaliana}, partial (40%)
Length = 627
Score = 39.3 bits (90), Expect = 0.002
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 6/80 (7%)
Frame = +3
Query: 218 KQRNSPLNSNWRGSRKTVP--WAWQTI---ISQPSQCPICYEDLDVTDSSF-LPCSCGFH 271
K+ L S G++K+ P ++W + S+ C +C E + D LPC+ FH
Sbjct: 315 KESIEELQSEVYGTKKSGPRKFSWSKLSWKASEQEDCAVCLESFKIGDKLIPLPCAHKFH 494
Query: 272 LCLFCHKKILEADARCPSCR 291
C K LE ++ CP CR
Sbjct: 495 ST--CLKPWLENNSHCPCCR 548
>BQ139229 weakly similar to GP|20453199|gb AT3g18290/MIE15_8 {Arabidopsis
thaliana}, partial (4%)
Length = 648
Score = 35.8 bits (81), Expect = 0.023
Identities = 14/46 (30%), Positives = 25/46 (53%)
Frame = +1
Query: 247 SQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRK 292
S CPIC + L + ++ + CG ++ C+ +E +CP C+K
Sbjct: 418 SGCPICTDYLFTSSAAVVSMPCGHYMHKQCYTLYMETAYKCPICKK 555
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 35.4 bits (80), Expect = 0.030
Identities = 32/128 (25%), Positives = 50/128 (39%), Gaps = 4/128 (3%)
Frame = +3
Query: 44 NVDSNGRMDNCPSSKHTAS--EENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTY 101
NV + +KH A EEN S +E+ +G D+E I ++D + DR
Sbjct: 522 NVHERREEQDEEENKHGAEVQEENESKSEEVEDEGGDVE---IDENDHEKSEADNDREDE 692
Query: 102 SHDESSGKDFSGSSSNSISTSCSGNDSEEEDDDGCLDDWE--AVADALYANDKSHSVVSE 159
DE K+ G D E+E+ G +++ E + Y D + S V+
Sbjct: 693 VVDEEKDKEEEGDDET------ENEDKEDEEKGGLVENHENHEAREEHYKADDASSAVAH 854
Query: 160 SPAEHEAE 167
E E
Sbjct: 855 DTHETSTE 878
>TC86451 similar to GP|21537318|gb|AAM61659.1 unknown {Arabidopsis
thaliana}, partial (51%)
Length = 995
Score = 35.4 bits (80), Expect = 0.030
Identities = 17/44 (38%), Positives = 22/44 (49%)
Frame = +2
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRK 292
CPIC + S+ CG C C K + A A+CP+CRK
Sbjct: 653 CPICMGPMVEEMST----RCGHIFCKSCIKAAISAQAKCPTCRK 772
>TC86450 similar to GP|21537318|gb|AAM61659.1 unknown {Arabidopsis
thaliana}, partial (39%)
Length = 1268
Score = 35.4 bits (80), Expect = 0.030
Identities = 17/44 (38%), Positives = 22/44 (49%)
Frame = +3
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRK 292
CPIC + S+ CG C C K + A A+CP+CRK
Sbjct: 741 CPICMGPMVEEMST----RCGHIFCKSCIKAAISAQAKCPTCRK 860
>TC83503 weakly similar to PIR|T51851|T51851 RING-H2 finger protein RHB1a
[imported] - Arabidopsis thaliana, partial (44%)
Length = 835
Score = 35.0 bits (79), Expect = 0.039
Identities = 17/49 (34%), Positives = 23/49 (46%)
Frame = +1
Query: 242 IISQPSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSC 290
+I + CPIC E+ DV + L C H L C + +E CP C
Sbjct: 568 VIEEEDGCPICLEEYDVENPKTL-SKCEHHFHLACILEWMERSDSCPIC 711
>BG645169 similar to PIR|D86157|D86 hypothetical protein AAF02892.1
[imported] - Arabidopsis thaliana, partial (6%)
Length = 779
Score = 34.3 bits (77), Expect = 0.066
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Frame = +1
Query: 266 CSCGFHLCLFCHKKIL--EADARCPSCRKLYDHV 297
C CGF +C C+K+I +CP C+ Y++V
Sbjct: 565 CECGFKVCRDCYKEICGNGGGGKCPGCKDSYNNV 666
>TC87578 similar to PIR|T12113|T12113 transcription factor - fava bean,
partial (49%)
Length = 1021
Score = 33.9 bits (76), Expect = 0.087
Identities = 35/137 (25%), Positives = 52/137 (37%), Gaps = 5/137 (3%)
Frame = +2
Query: 4 GFISDPPLS-----KKKRTNGSAKLKQIKLDVRREQWLSRVKKGCNVDSNGRMDNCPSSK 58
GFIS L + T G AK+ + D + L R++ D +
Sbjct: 386 GFISSKGLKIMNLGDAQPTTGVAKVLEGDDDDAVDPHLERIRNEAGEDES---------- 535
Query: 59 HTASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSSSNS 118
EE+ E +G + + DSD+ S D + KD S +S S
Sbjct: 536 ---DEEDEDFVAEKDDEGSPTDDSGADDSDASQ---SGDEKEIPAKKEPKKDLSSKASAS 697
Query: 119 ISTSCSGNDSEEEDDDG 135
STS S S++ D+DG
Sbjct: 698 TSTSTSKKKSKDADEDG 748
>AW584214 similar to GP|7363283|db Similar to Arabidopsis thaliana DNA
chromosome 4 BAC clone F20D10; putative protein.
(AL035538), partial (9%)
Length = 626
Score = 33.9 bits (76), Expect = 0.087
Identities = 13/32 (40%), Positives = 17/32 (52%), Gaps = 1/32 (3%)
Frame = +3
Query: 264 LPCSCGFHLCLFCHKKIL-EADARCPSCRKLY 294
LPC C F +C C+K L + CP C + Y
Sbjct: 414 LPCECDFKICRNCYKDTLRNGEGVCPGCNEAY 509
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 33.5 bits (75), Expect = 0.11
Identities = 26/92 (28%), Positives = 36/92 (38%)
Frame = -3
Query: 80 EGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSSSNSISTSCSGNDSEEEDDDGCLDD 139
E C R I S S+ S SS S SSS+S S+SCS + S
Sbjct: 301 ESICNNSGSGRVLILSISSSSPSSSSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSS---- 134
Query: 140 WEAVADALYANDKSHSVVSESPAEHEAECRYT 171
+ + ++ S S S SP+ + C T
Sbjct: 133 -SSSCSSSSSSSCSSSSSSTSPSSSSSLCSST 41
>TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.7
[imported] - Arabidopsis thaliana, partial (64%)
Length = 1328
Score = 33.5 bits (75), Expect = 0.11
Identities = 33/117 (28%), Positives = 46/117 (39%), Gaps = 14/117 (11%)
Frame = -3
Query: 21 AKLKQIKLDVRREQWLSRV--KKGCNVDSNGRMDNCPSSKHTASEENRSSCKEMTRKGED 78
A + LD R W K G N++S S TA + + SS D
Sbjct: 378 AMASSLVLDSRERDWNLET*SKPGTNLNS--------CSMKTAIDSSSSSANN------D 241
Query: 79 IEGTCIQDSDSR------------STINSPDRSTYSHDESSGKDFSGSSSNSISTSC 123
+G C D +SR S +SP RS+ S SG +G+ S+ I+ SC
Sbjct: 240 DDGNCDDDDESRSEEHSGKSPVRTSDESSPQRSSQSEVLDSGMRIAGNCSSIIALSC 70
>TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidopsis
thaliana}, partial (6%)
Length = 890
Score = 33.1 bits (74), Expect = 0.15
Identities = 21/61 (34%), Positives = 32/61 (52%), Gaps = 1/61 (1%)
Frame = +1
Query: 82 TCIQDSDSRSTINSPDRSTYSHDESSGKDF-SGSSSNSISTSCSGNDSEEEDDDGCLDDW 140
T D S + D+ S ES + S S S++ S+S SG+DS+E+D+D D+
Sbjct: 361 TLTGDPPSVEVNGNEDKDDGSEGESESESSESESESSTSSSSTSGSDSDEDDEDDAEDEE 540
Query: 141 E 141
E
Sbjct: 541 E 543
>TC82350 similar to GP|9837385|gb|AAG00554.1| retinitis pigmentosa GTPase
regulator-like protein {Takifugu rubripes}, partial (2%)
Length = 759
Score = 32.7 bits (73), Expect = 0.19
Identities = 25/111 (22%), Positives = 48/111 (42%)
Frame = +1
Query: 67 SSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSSSNSISTSCSGN 126
S +E+ +K E+ ++DSD N+ + + Y +E+ G++ +S N N
Sbjct: 40 SRMEEINQKEEEDHPRAVEDSD-----NANEDTQYGVEET-GEEEEDASENEEEEEEEEN 201
Query: 127 DSEEEDDDGCLDDWEAVADALYANDKSHSVVSESPAEHEAECRYTVLEDDK 177
D+ +EDD D + ND ++ + H+ Y L++ K
Sbjct: 202 DTVDEDDQFIFGAGVNPLDFVRNNDSGVNLYQKFKDYHQKSIEYRALDNRK 354
>AW696663 weakly similar to GP|17827193|dbj ORF1 {TT virus}, partial (2%)
Length = 660
Score = 32.0 bits (71), Expect = 0.33
Identities = 16/34 (47%), Positives = 20/34 (58%)
Frame = +1
Query: 44 NVDSNGRMDNCPSSKHTASEENRSSCKEMTRKGE 77
NV NGR+ N PSS + A+ E SS R+GE
Sbjct: 349 NVVDNGRLQNFPSSSYEAAMEKLSSLITRQRRGE 450
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.129 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,906,142
Number of Sequences: 36976
Number of extensions: 192049
Number of successful extensions: 1157
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 1115
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1129
length of query: 322
length of database: 9,014,727
effective HSP length: 96
effective length of query: 226
effective length of database: 5,465,031
effective search space: 1235097006
effective search space used: 1235097006
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0081b.3


