

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0079c.8
(757 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
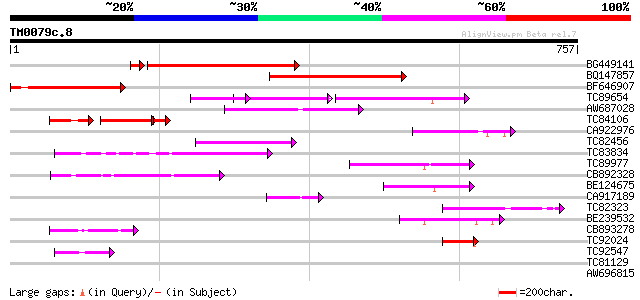
Score E
Sequences producing significant alignments: (bits) Value
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 254 1e-70
BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {A... 189 4e-48
BF646907 similar to GP|15983442|gb| At1g10240/F14N23_12 {Arabido... 172 4e-43
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 89 2e-31
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 127 2e-29
TC84106 similar to GP|9502168|gb|AAF88018.1| contains simlarity ... 77 8e-26
CA922976 weakly similar to GP|9502168|gb|A contains simlarity to... 114 1e-25
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 108 1e-23
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 90 4e-18
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 87 3e-17
CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sin... 77 2e-14
BE124675 weakly similar to GP|13365573|db hypothetical protein~s... 59 5e-09
CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.3... 54 2e-07
TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea ma... 54 2e-07
BE239532 53 5e-07
CB893278 weakly similar to PIR|T05645|T056 hypothetical protein ... 50 4e-06
TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10... 49 7e-06
TC92547 similar to PIR|A56235|A56235 transcription activator Maf... 44 2e-04
TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 37 0.028
AW696815 homologue to GP|11994736|dbj far-red impaired response ... 33 0.31
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 254 bits (649), Expect(2) = 1e-70
Identities = 109/203 (53%), Positives = 161/203 (78%)
Frame = +3
Query: 184 DQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIRNFIQSQSSIGKESDASNVL 243
D++RI++ +K G S+ ++R++ELEK ++ LPF E+D+RN +QS + E + ++L
Sbjct: 69 DKNRILMFAKTGISVHQMMRLMELEKCVEPGYLPFTEKDVRNLLQSFRKLDPEEETLDLL 248
Query: 244 KLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMP 303
++C+++KD+D +FK+++TLD NN+LE+I W++ SI+ Y+ FGD VVFDTT+R+ +DMP
Sbjct: 249 RMCRNIKDKDPNFKFEYTLDANNRLENIAWSYASSIQLYDIFGDAVVFDTTHRLTAFDMP 428
Query: 304 LGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAI 363
LG+WVG++N+G FFGCVLLRDE + SFSWA+K+FL F+NGK P+TILTDQ++ LKEA+
Sbjct: 429 LGLWVGINNYGMPCFFGCVLLRDETVRSFSWAIKAFLGFMNGKAPQTILTDQNICLKEAL 608
Query: 364 AMELPNTKHAFCIWHIVAKLSSW 386
+ E+P TKHAFCIW IVAK SW
Sbjct: 609 SAEMPMTKHAFCIWMIVAKFPSW 677
Score = 31.6 bits (70), Expect(2) = 1e-70
Identities = 12/19 (63%), Positives = 17/19 (89%)
Frame = +2
Query: 162 NHQLLDDKEVQFLPAYRSI 180
NH+LL+ +V+FLPAYR+I
Sbjct: 2 NHELLEPNQVRFLPAYRTI 58
>BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (26%)
Length = 560
Score = 189 bits (479), Expect = 4e-48
Identities = 87/182 (47%), Positives = 130/182 (70%)
Frame = +1
Query: 348 PKTILTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEFHKVYH 407
P+T+LTD + LKEAIA+E+P +KHAFCIWHI++K S W LGS+Y+++K +FH++Y+
Sbjct: 4 PQTLLTDHNTWLKEAIAVEMPESKHAFCIWHILSKFSDWXYLLLGSQYDEWKADFHRLYN 183
Query: 408 LECEDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRSESINS 467
LE +DFE W M+ ++GL +KHI LYS R FWAL +L+ +FFAG+T+ ++ESIN
Sbjct: 184 LEMXEDFEKSWRQMVDKYGLHANKHIISLYSLRTFWALPFLRRYFFAGLTSXCQTESINV 363
Query: 468 YIKRFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRTGFPIEEQAASMLTP 527
+I+RFL ++ F+ QV V+ ++AG + M++K ++TG PIE AA++LTP
Sbjct: 364 FIQRFLSAQSQPXRFLQQVADIVDFNDRAGAKQXMQRKMQKXCLKTGSPIESHAATILTP 543
Query: 528 YA 529
YA
Sbjct: 544 YA 549
>BF646907 similar to GP|15983442|gb| At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (4%)
Length = 621
Score = 172 bits (436), Expect = 4e-43
Identities = 88/155 (56%), Positives = 110/155 (70%), Gaps = 1/155 (0%)
Frame = +1
Query: 1 MTSEEENDVTDWPLELFKDHASMCEDNGNTNS-STCQEPQLEPLSLANNSADWIPYEGQI 59
M+SE N+ D LE ED+ +S +T EPQ PL AN+ D + YEG++
Sbjct: 181 MSSEGRNEAIDGSLEY--------EDDEIIDSLTTVPEPQHGPLPQANHIDDGVLYEGKV 336
Query: 60 FNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGITKQSKV 119
FNS+ +AY+FYCLFARK+GFSIRR H+YKS KNQS++NPLG+YKREFVCHRAG K
Sbjct: 337 FNSDDEAYNFYCLFARKNGFSIRRHHVYKSIKNQSDDNPLGVYKREFVCHRAGTISVDKD 516
Query: 120 SEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVV 154
+EVE +RKRK+SRCNCGAK+LV TI +KWVV
Sbjct: 517 NEVEGKRKRKSSRCNCGAKLLVNITTINSXKKWVV 621
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 89.4 bits (220), Expect(2) = 2e-31
Identities = 54/183 (29%), Positives = 88/183 (47%), Gaps = 4/183 (2%)
Frame = +1
Query: 436 LYSNRQFWALAYLKDFFFAGMTTTGRSESINSYIKRFLHVRTSLTDFVNQVGIAVNIRNQ 495
+Y+ RQ W YL+ FF + +E +N + ++ T+L V Q AV+ ++
Sbjct: 715 MYNARQHWVPVYLRGSFFGEIPLNDGNECLNFFFDGHVNASTTLQLLVRQYEKAVSTWHE 894
Query: 496 AGEEARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHEIELSTKYASTEEGENSYIV 555
+A NP +RT P+E+QAAS+ T F Q E+ + +T+ ++ I
Sbjct: 895 RELKADFETSNSNPVLRTPSPMEKQAASLYTRKMFMKFQEELVGTMANPATKIEDSGTIT 1074
Query: 556 RFHTKVDG----GRIVKWIEEDKSINCSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLP 611
+ G V + + +CSC+ FE+SGI+CRH + V KN LP+ Y+
Sbjct: 1075TYRVAKFGENQTSHTVAFNSSEMKASCSCQMFEYSGIVCRHILAVFRAKNVLTLPSHYVL 1254
Query: 612 FRW 614
RW
Sbjct: 1255KRW 1263
Score = 65.9 bits (159), Expect(2) = 2e-31
Identities = 43/133 (32%), Positives = 68/133 (50%), Gaps = 1/133 (0%)
Frame = +3
Query: 300 YDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVAL 359
Y +PL + G+ N + +L+ +E PS+ W K++L V+G+ P +I TD D +
Sbjct: 312 YHLPLSL--GLIIMVNLFYLAVLLILNESEPSYIWLFKTWLRAVSGRPPVSITTDLDPVI 485
Query: 360 KEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEFHKVYHLECE-DDFEHKW 418
+ A+A LP T+H FC W I + S + S F EF K H D+FE W
Sbjct: 486 QVAVAQVLPPTRHRFCKWSIFRENRSKLAHLYQSN-PTFDNEFKKCVHESVTIDEFESCW 662
Query: 419 TLMIAQFGLGMDK 431
++ ++ + MDK
Sbjct: 663 RSLLERYYI-MDK 698
Score = 62.4 bits (150), Expect = 6e-10
Identities = 29/80 (36%), Positives = 43/80 (53%)
Frame = +2
Query: 242 VLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYD 301
VL K ++ + +F Y D +N +I WA Y FGD V+FDTTY+ N+Y
Sbjct: 131 VLDYLKRMRAENHAFFYAVQSDVDNAGGNIFWADETCRTNYSYFGDTVIFDTTYKTNQYR 310
Query: 302 MPLGIWVGVDNHGNSIFFGC 321
+P + G ++HG + FGC
Sbjct: 311 VPFASFTGFNHHGQPVLFGC 370
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 127 bits (318), Expect = 2e-29
Identities = 66/187 (35%), Positives = 100/187 (53%), Gaps = 1/187 (0%)
Frame = +1
Query: 287 DVVVFDTTYRINRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGK 346
DV+ FDTTY+ N+Y+ PL I+ G ++H +I FG LL DE I S+ W L FL + K
Sbjct: 1 DVLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENK 180
Query: 347 HPKTILTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEFHK-V 405
PK ++TD D +++EAI P+ H C WH+ + F F K +
Sbjct: 181 FPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQE------NIKKTPFLEGFRKAM 342
Query: 406 YHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRSESI 465
Y + FE W+ +I + L + + Y+N+ WA AYL+D FF + TT + E+I
Sbjct: 343 YSNFTPEQFEDFWSELIQKNELEGNAWVIKTYANKSLWATAYLRDKFFGRIRTTSQCEAI 522
Query: 466 NSYIKRF 472
N+ +K +
Sbjct: 523 NAIVKTY 543
>TC84106 similar to GP|9502168|gb|AAF88018.1| contains simlarity to
Arabidopsis thaliana far-red impaired response protein
(GB:AAD51282.1), partial (25%)
Length = 673
Score = 77.4 bits (189), Expect(3) = 8e-26
Identities = 37/74 (50%), Positives = 48/74 (64%)
Frame = +2
Query: 122 VESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEVQFLPAYRSIP 181
V R RK+ RC C AKM ++ + +W V F N HNH+LL+D +V+ LPAYR I
Sbjct: 383 VMHHRDRKSVRCGCDAKMYLSKDVVDGVSQWTVLQFSNVHNHELLEDDQVRLLPAYRKIH 562
Query: 182 IVDQDRIMLLSKAG 195
DQ+RI+LLSKAG
Sbjct: 563 EADQERILLLSKAG 604
Score = 55.1 bits (131), Expect(3) = 8e-26
Identities = 28/60 (46%), Positives = 40/60 (66%), Gaps = 1/60 (1%)
Frame = +1
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNP-LGIYKREFVCHRAG 112
PY G +F S+ +A++ Y FARK GFS+R K +S +P LG+YKR+FVC+R+G
Sbjct: 199 PYVGMVFKSDDEAFEHYGNFARKHGFSMR--------KARSRISPQLGVYKRDFVCYRSG 354
Score = 23.5 bits (49), Expect(3) = 8e-26
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 2/24 (8%)
Frame = +3
Query: 193 KAGCSISLIIRVLELEKGI--DTC 214
K G + I+++LELEKGI D C
Sbjct: 597 KLGFRYTRIVKMLELEKGIQGDIC 668
>CA922976 weakly similar to GP|9502168|gb|A contains simlarity to Arabidopsis
thaliana far-red impaired response protein
(GB:AAD51282.1), partial (11%)
Length = 827
Score = 114 bits (285), Expect = 1e-25
Identities = 65/148 (43%), Positives = 88/148 (58%), Gaps = 11/148 (7%)
Frame = -3
Query: 539 LSTKYASTEEGENSYIVRFHTKVDGGRIVKWIEEDKSINCSCKEFEFSGILCRHAIRVLV 598
L+ +YA++E SYI+R K+DG R V W+ E + I CSCKEFE SGILCRHA+R+LV
Sbjct: 819 LTMQYATSELYNGSYIIRHSKKLDGERHVLWLPEKEQILCSCKEFESSGILCRHALRILV 640
Query: 599 MKNYFHLPTKYLPFRWRQESSLVPTSSHIINCNDGSSF-------EFRSLVQWLQVESLK 651
KNYF LP KY RW +E SL+ H + SF E++S + L ES
Sbjct: 639 TKNYFQLPEKYYLSRWHRERSLIVEDEH-----NNQSFIAEKWFQEYQSQAENLFSESSI 475
Query: 652 TKDREEVA----IRELERVIRIITDMPE 675
TK+R + A ++EL R++ + DM E
Sbjct: 474 TKERSDYARKELMKELTRILNEVRDMSE 391
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 108 bits (269), Expect = 1e-23
Identities = 45/135 (33%), Positives = 81/135 (59%)
Frame = +2
Query: 249 LKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWV 308
++ ++ F +D+ ++++ WA AYE FG+V+ DTTY ++YD+PL +V
Sbjct: 749 MQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDTTYLTSKYDLPLVPFV 928
Query: 309 GVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELP 368
GV++HG +I GC +L + + +W +L ++G P I+T++D A+K AI + P
Sbjct: 929 GVNHHGQTILLGCAILSNLDAKTLTWLFTRWLECMHGHAPNGIITEEDKAMKNAIEVAFP 1108
Query: 369 NTKHAFCIWHIVAKL 383
+H +C+WHI+ K+
Sbjct: 1109KARHRWCLWHIMKKV 1153
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 89.7 bits (221), Expect = 4e-18
Identities = 70/293 (23%), Positives = 129/293 (43%), Gaps = 1/293 (0%)
Frame = +3
Query: 60 FNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGITKQSKV 119
F S DAY +Y +A++ GF +R + + ++ + Y C G + V
Sbjct: 231 FESYDDAYSYYICYAKEVGFCVRVKNSWFKRNSKEK------YGAVLCCSSQGFKRTKDV 392
Query: 120 SEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEVQFLPAYRS 179
+ + RK +R C A + + + ++W + HNH L ++ S
Sbjct: 393 NNL-----RKETRTGCPAMIRMK---LVESQRWRICEVTLEHNHVL--GAKIHKSIKKNS 542
Query: 180 IPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIRNFIQSQSSIG-KESD 238
+P D + G +I + ++ +G D L RD R F + + + ++ D
Sbjct: 543 LPSSDAE--------GKTIKVYHALVIDTEGNDN--LNSNARDDRAFSKYSNKLNLRKGD 692
Query: 239 ASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRIN 298
+ ++ + +F Y ++ L + +W S A F DV+ FD TY +N
Sbjct: 693 TQAIYNFLCRMQLTNPNFFYLMDFNDEGHLRNALWVDAKSRAACGYFSDVIYFDNTYLVN 872
Query: 299 RYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPKTI 351
+Y++PL VG+++HG S+ GC LL E I S+ W ++++ + P+TI
Sbjct: 873 KYEIPLVALVGINHHGQSVLLGCGLLAGEIIESYKWLFRTWIKCIPRCSPQTI 1031
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 86.7 bits (213), Expect = 3e-17
Identities = 52/171 (30%), Positives = 92/171 (53%), Gaps = 4/171 (2%)
Frame = +2
Query: 454 AGMTTTGRSESINSYIKRFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRT 513
AG +T RSES+NS+ +++H + +L +FV Q + + R E A + P +++
Sbjct: 2 AGTSTAQRSESMNSFFDKYIHKKITLKEFVKQYRLILLNRYDEEEIADFDTLHKQPALKS 181
Query: 514 GFPIEEQAASMLTPYAFKLLQHEI-ELSTKYASTEEGENS---YIVRFHTKVDGGRIVKW 569
P E+Q +++ T FK Q E+ ++ + E G+ + YIV+ + K D +V W
Sbjct: 182 PSPWEKQMSTIYTHAIFKKFQVEVLGVAGCQSRIEVGDGTAVRYIVQDYEK-DEEFLVTW 358
Query: 570 IEEDKSINCSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQESSL 620
E ++C C+ FE+ G LCRHA+ VL +P+ Y+ RW +++ +
Sbjct: 359 KELSSEVSCFCRLFEYKGFLCRHALSVLQRCGCSSVPSHYIMKRWTKDAKI 511
>CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sinorhizobium
meliloti}, partial (1%)
Length = 828
Score = 77.0 bits (188), Expect = 2e-14
Identities = 62/234 (26%), Positives = 107/234 (45%), Gaps = 2/234 (0%)
Frame = +1
Query: 55 YEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGIT 114
Y GQ+ NS+ +AYD Y A K+GFS+R K + +N ++F C + G
Sbjct: 157 YLGQVVNSKDEAYDLYQEHAFKTGFSVR-----KGKELYYDNEKKKTRLKDFYCSKQGFK 321
Query: 115 KQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEVQFL 174
EV KR SR NC + + + E W V HNH+ + ++ L
Sbjct: 322 NNEPDGEV--AYKRADSRTNC---LAMVRFNVTKEGVWKVTKLILDHNHEFVPLQQRYLL 486
Query: 175 PAYRSIPIVDQDRIMLLSKAGCSISLIIRVLELE-KGIDTCTLPFLERDIRNFIQS-QSS 232
+ R++ + +DRI L ++ + L E G+D + + +D+ N++ + +
Sbjct: 487 RSMRNMSNLKEDRIKSLVNDAIRVTNVGGYLGKEVGGVDKRGV--MLKDMHNYVSTGKLK 660
Query: 233 IGKESDASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFG 286
+ DA ++L +S + +D F Y LD ++L ++ W G S+ Y A G
Sbjct: 661 FIEAGDAQSLLNHLQSKQAQDSMFYYSVQLDHESRLNNVFWRDGKSVVDYIATG 822
>BE124675 weakly similar to GP|13365573|db hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (4%)
Length = 448
Score = 59.3 bits (142), Expect = 5e-09
Identities = 34/125 (27%), Positives = 58/125 (46%), Gaps = 3/125 (2%)
Frame = +2
Query: 499 EARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHEIELSTKYASTEEGENSYIVRFH 558
EA K P + + + A+ TP AF++ Q E S + + Y+ +
Sbjct: 41 EASDEMKGCLPKLMGNVVVLKHASVAYTPRAFEVFQQRYEKSLNVIVNQHKRDGYLFEYK 220
Query: 559 TKVDGGR---IVKWIEEDKSINCSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWR 615
G V + D ++ CSC +FE G LC HA++VL +N +P++Y+ RW
Sbjct: 221 VNTYGHARQYTVTFSSSDNTVVCSCMKFEHVGFLCSHALKVLDNRNIKVVPSRYILKRWT 400
Query: 616 QESSL 620
+++ L
Sbjct: 401 KDARL 415
>CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (12%)
Length = 312
Score = 53.9 bits (128), Expect = 2e-07
Identities = 27/78 (34%), Positives = 45/78 (57%), Gaps = 1/78 (1%)
Frame = +1
Query: 343 VNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEF 402
++G+ P +I TD D ++ AI P+T+H FC WHI + S + ++ +F+ EF
Sbjct: 10 MSGRPPLSITTDHDSVIQSAIMQVFPDTRHRFCKWHIFKQCQEKLS-HIFLQFPNFEAEF 186
Query: 403 HKVYHL-ECEDDFEHKWT 419
HK +L + D+FE W+
Sbjct: 187 HKCVNLTDSIDEFESCWS 240
>TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea mays},
partial (2%)
Length = 1165
Score = 53.9 bits (128), Expect = 2e-07
Identities = 44/167 (26%), Positives = 71/167 (42%), Gaps = 4/167 (2%)
Frame = +1
Query: 578 CSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQESSLVPTSSHIINCNDGSSFE 637
CSC+ FE GI C H V+ ++ H+P + RW + + +S D + E
Sbjct: 31 CSCRLFESRGIPCCHVFFVMKEEDVDHIPKCLVMTRWTKNAKGEFLNSDSNGEIDANMIE 210
Query: 638 FRSLVQWLQVES--LKTKDREEVAIRELERVIRIITDMPEEQEHAVDMEPNVPNSN--DC 693
+ + K ++ R+ I+ ++ Q+ D+E + + + D
Sbjct: 211 LARFGAYCSAFTTFCKEASKKNGVFRD------IMDEILNLQKKYCDVEDSTRSKSFTDD 372
Query: 694 DVGNPIISKSKGRPKGSRPKGGVEVAKKPRHCHVPNCGGTDHDARNC 740
VG+P+ KSKG PK + AK RHC C T H AR+C
Sbjct: 373 QVGDPVTVKSKGAPKKKK-----NAAKSVRHC--SRCKSTSHTARHC 492
>BE239532
Length = 645
Score = 52.8 bits (125), Expect = 5e-07
Identities = 39/161 (24%), Positives = 77/161 (47%), Gaps = 21/161 (13%)
Frame = +3
Query: 521 AASMLTPYAFKLLQHEIELSTKYASTEEGENS-----YIVRFHTKVDGGRIVKWIEEDKS 575
A+++ T F++ +E T +++ E S + V + K + V + +
Sbjct: 54 ASTVYTHKGFRIFLNEYLEGTGGSTSIEISQSNNLSNHEVMLNHKPNKKYAVTFDSSNMK 233
Query: 576 INCSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQESSLV------------PT 623
INCSC++F+ GILC HA+R+ +K +P +Y RW + + V T
Sbjct: 234 INCSCRKFDSMGILCSHALRIYNIKGILRIPDQYFLKRWSKNARSVIYEHMPRGMGEDST 413
Query: 624 SSHIINCNDGSSFEFRSLVQ----WLQVESLKTKDREEVAI 660
S+ +IN +D + +R+ + +L +ES +K+ + + +
Sbjct: 414 SNSVIN-DDDAGILYRNAIMTSFYYLVLESQDSKEAQNIYV 533
>CB893278 weakly similar to PIR|T05645|T056 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (5%)
Length = 821
Score = 49.7 bits (117), Expect = 4e-06
Identities = 37/119 (31%), Positives = 55/119 (46%), Gaps = 1/119 (0%)
Frame = +3
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGI 113
P+ G FNS +A FY + R+ GF++R H +S N N +G ++FVC + G
Sbjct: 315 PFIGMQFNSREEARGFYDGYGRRIGFTVRIHHNRRSRVN---NELIG---QDFVCSKEGF 476
Query: 114 TKQSKVSEVES-QRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEV 171
+ V + +R C A + + R E KWVV F H H+L+ EV
Sbjct: 477 RAKKYVHRKDRVLPPPPATREGCQAMIRLALRD---EGKWVVTKFVKEHTHKLMSPGEV 644
>TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (11%)
Length = 938
Score = 48.9 bits (115), Expect = 7e-06
Identities = 24/53 (45%), Positives = 32/53 (60%), Gaps = 4/53 (7%)
Frame = +3
Query: 578 CSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQES----SLVPTSSH 626
CSC+ FEFSG+LCRH + V + N LP+ Y+ RW + + SL SSH
Sbjct: 93 CSCQMFEFSGLLCRHILAVFRVTNVLTLPSHYILKRWTKNAKSNVSLQEHSSH 251
>TC92547 similar to PIR|A56235|A56235 transcription activator MafB - chicken,
partial (6%)
Length = 1158
Score = 44.3 bits (103), Expect = 2e-04
Identities = 24/81 (29%), Positives = 44/81 (53%), Gaps = 1/81 (1%)
Frame = +3
Query: 60 FNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGITKQSKV 119
F +AY+FY + + GFSIR+ + +++ I REFVC++ G+ + +
Sbjct: 936 FGPADEAYEFYYRYGKCKGFSIRKGDVRRNSSGI-------ITMREFVCNKNGLRDKKHL 1094
Query: 120 SEVESQR-KRKTSRCNCGAKM 139
S + +R R+ +R NC A++
Sbjct: 1095 SRNDRKRDHRRLTRTNCEARL 1157
>TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (13%)
Length = 721
Score = 37.0 bits (84), Expect = 0.028
Identities = 31/118 (26%), Positives = 48/118 (40%), Gaps = 2/118 (1%)
Frame = +2
Query: 43 LSLANNSADWIPYEGQIFNSEGDAYDFYCLFARKSGF--SIRRDHIYKSTKNQSENNPLG 100
++ D+ P G F + AY FY +A+ GF SI+ K TK
Sbjct: 380 IAFFEGDTDFEPPNGIEFETHEAAYSFYQEYAKSMGFTTSIKNSRRSKKTKE-------- 535
Query: 101 IYKREFVCHRAGITKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFH 158
+F C R G+T + S+ S ++ + +C A M V G KW + F+
Sbjct: 536 FIDAKFACSRYGVTPE---SDGRSSQRSSVKKTDCKACMHVKRNPDG---KWNIFEFY 691
>AW696815 homologue to GP|11994736|dbj far-red impaired response protein;
Mutator-like transposase-like protein; phytochrome A
signaling, partial (7%)
Length = 394
Score = 33.5 bits (75), Expect = 0.31
Identities = 22/91 (24%), Positives = 38/91 (41%)
Frame = +2
Query: 26 DNGNTNSSTCQEPQLEPLSLANNSADWIPYEGQIFNSEGDAYDFYCLFARKSGFSIRRDH 85
+ G+ NS T +++ + P G F S G+AY FY +AR GF+ +
Sbjct: 149 NGGDLNSPTVD------IAMFKEDTNLEPLSGMEFESHGEAYSFYQEYARSMGFNTAIQN 310
Query: 86 IYKSTKNQSENNPLGIYKREFVCHRAGITKQ 116
+S ++ +F C R G ++
Sbjct: 311 SRRSKTSRE------FIDAKFACSRYGTKRE 385
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,977,331
Number of Sequences: 36976
Number of extensions: 429741
Number of successful extensions: 2266
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 2216
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2252
length of query: 757
length of database: 9,014,727
effective HSP length: 103
effective length of query: 654
effective length of database: 5,206,199
effective search space: 3404854146
effective search space used: 3404854146
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0079c.8


