

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0079b.1
(1571 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
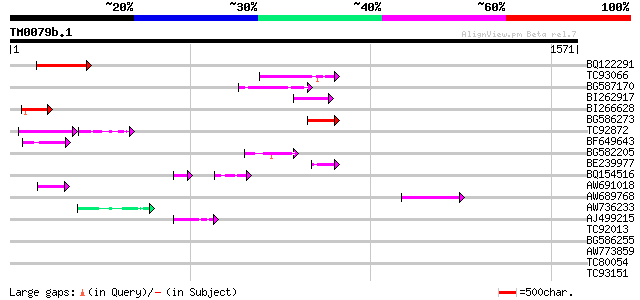
Score E
Sequences producing significant alignments: (bits) Value
BQ122291 127 3e-29
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 125 1e-28
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 119 7e-27
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 82 2e-15
BI266628 79 2e-14
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 69 1e-11
TC92872 51 8e-11
BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element pol... 55 2e-07
BG582205 54 4e-07
BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryz... 52 1e-06
BQ154516 52 2e-06
AW691018 weakly similar to GP|2462935|em open reading frame 1 {B... 51 3e-06
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 49 2e-05
AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Dani... 47 8e-05
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 43 8e-04
TC92013 homologue to GP|11595557|emb|CAC18142. related to c-modu... 43 0.001
BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l ... 42 0.002
AW773859 41 0.003
TC80054 SP|Q28009|FUS_BOVIN RNA-binding protein FUS (Pigpen prot... 41 0.004
TC93151 homologue to GP|22136928|gb|AAM91808.1 unknown protein {... 41 0.004
>BQ122291
Length = 487
Score = 127 bits (320), Expect = 3e-29
Identities = 58/153 (37%), Positives = 96/153 (61%)
Frame = -2
Query: 74 DEDRANSVENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHS 133
D R NPA+ WE DS L TW+LS +SPS+L V +HSWQ+W+ + +
Sbjct: 486 DAAREAGTRNPAYTEWEDKDSLLCTWILSRISPSLLSRFVLLRHSWQVWDEIHSYCFTQM 307
Query: 134 LAQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYS 193
+S QLRSELRSITKGS+ S+++ RI++I +L SIGDPV++R+ +E + + L E++
Sbjct: 306 KTRSRQLRSELRSITKGSRTVSEFIARIRAISESLASIGDPVSHRDLIEVVLEALPEEFD 127
Query: 194 HLMTIVTSREFPCSISQAEAMVIAHEARLDRLR 226
++ V ++ S+ + E+ ++ E+R ++ +
Sbjct: 126 PIVASVNAKSEVVSLDELESQLLTQESRKEKFK 28
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 125 bits (315), Expect = 1e-28
Identities = 83/238 (34%), Positives = 116/238 (47%), Gaps = 15/238 (6%)
Frame = +1
Query: 692 LELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKF 751
L+ I +DLWGP+ + S G Y +T +D F R W+Y L+ K+ET F +++ +VE +
Sbjct: 97 LDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQT 276
Query: 752 GLKIKSVQTDGGTEF--KPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLL 809
G +K + TD EF GI T P QNG ER R ++E +L
Sbjct: 277 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 456
Query: 810 ACANMPT--SYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVP--------ADYKILRV 859
+ A + W A TA L+NR P L FKVP DY LR+
Sbjct: 457 SNAGL*N*RDLWVEAASTACHLVNRSPHSALD--------FKVPEDIWSGNLVDYSNLRI 612
Query: 860 FGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGYKCLASD---GRIYISKDVQFNE 914
FG Y + N KL+ R+ EC+FL Y+S KGY+ SD ++ +S+DV FNE
Sbjct: 613 FGCPAYALV---NDGKLAPRAGECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNE 777
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 119 bits (299), Expect = 7e-27
Identities = 76/209 (36%), Positives = 110/209 (52%), Gaps = 5/209 (2%)
Frame = -3
Query: 635 LWHSRLGHPHYETLQSALTTCKISVPHKSKYTICHSCCVAKSHRLPSSASSTLYTAPLEL 694
LWH+RLGHPH L L V ++K C +C + K + +ST+Y +L
Sbjct: 584 LWHARLGHPHGRALNLMLP----GVVFENKN--CEACILGKHCKNVFPRTSTVYENCFDL 423
Query: 695 IFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLK 754
I+ DLW S+ S + YF+T +D S++TW+ L+ K F FQA V + K
Sbjct: 422 IYTDLWTAPSL-SRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAK 246
Query: 755 IKSVQTDGGTE-----FKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLL 809
IK +++D G E FK L H GI H+ +CP+T QNG +RK++H++E SL+
Sbjct: 245 IKILRSDNGGEYTSYAFKSHLDHH---GILHQTSCPYTPQQNGVAKRKNKHLMEVARSLM 75
Query: 810 ACANMPTSYWDHAFLTATFLINRLPTPVL 838
AN+ TA +LIN +PT VL
Sbjct: 74 FQANVS---------TACYLINWIPTKVL 15
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 81.6 bits (200), Expect = 2e-15
Identities = 42/110 (38%), Positives = 60/110 (54%)
Frame = +1
Query: 786 HTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYH 845
HT QNG ER +R ++E ++L A M S+W A TA ++INR P+ V+ +P
Sbjct: 82 HTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPME 261
Query: 846 KLFKVPADYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGY 895
P DY L VFG Y +KL +S++C+FLGY+ + KGY
Sbjct: 262 MWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGY 411
>BI266628
Length = 536
Score = 78.6 bits (192), Expect = 2e-14
Identities = 39/95 (41%), Positives = 59/95 (62%), Gaps = 7/95 (7%)
Frame = -1
Query: 32 LLLKLDDSNF-------LSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRANSVENP 84
+++K+D SN L WKQ VEG++R + F++ P+IPP +L+DE R + P
Sbjct: 455 VVIKVDPSNKQVLS*VRLQWKQQVEGVLRETKTVKFVI-SPQIPPVYLTDEAREAGTKKP 279
Query: 85 AFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSW 119
A+ WE+ DS L TW+LS++ PS+L V +HSW
Sbjct: 278 AYTEWEEQDSLLCTWILSTI*PSLLSRFVLLRHSW 174
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 69.3 bits (168), Expect = 1e-11
Identities = 36/90 (40%), Positives = 56/90 (62%), Gaps = 1/90 (1%)
Frame = -2
Query: 826 ATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSSKLSFRSQECVF 885
A +LINR+PT VL + +P+ L + +RVFG CY + +KL RS++ +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 886 LGYSSSHKGYKCLASDG-RIYISKDVQFNE 914
+GYS++ KGYKC + R+ +S+DV+F E
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIE 435
>TC92872
Length = 923
Score = 50.8 bits (120), Expect(2) = 8e-11
Identities = 37/165 (22%), Positives = 74/165 (44%)
Frame = +1
Query: 24 SSTSLVHKLLLKLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRANSVEN 83
SS L + KL NF +WK+ V ++ + + I + + S N
Sbjct: 73 SSLILNASITFKLSRKNFRAWKRQVTTLLAGIEVMGHIDGTTPIQTETIINN--GVSAPN 246
Query: 84 PAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSE 143
P + W D + LLSS++ + + + + + +W A+ F+ S + + ++
Sbjct: 247 PDYTKWFTLDQLIINLLLSSMTEADSISFASYETARTLWVAIESQFNNTSRSHVMSVTNQ 426
Query: 144 LRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGL 188
++ TKG K+ +DYL +KS+ + L I ++ + + +GL
Sbjct: 427 IQRCTKGDKSITDYLFSVKSLADELAVIDKSLSDDDITLFVLNGL 561
Score = 35.4 bits (80), Expect(2) = 8e-11
Identities = 35/155 (22%), Positives = 64/155 (40%), Gaps = 1/155 (0%)
Frame = +2
Query: 191 DYSHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTP 250
+Y+ + + +R+ P + + + ++AH+ L R + Q P+A A T+P Q +
Sbjct: 569 EYNDIAASIRTRQHPFTFEELHSHLLAHDDYLRREAV-QVDIQVPTANFAHHTSPNQRSS 745
Query: 251 QAPPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGG 310
+ ++P P + N S+ +S+ + RG
Sbjct: 746 KDDGILP---------------IPSTGRGNNFSK--------------QSQHHGSNTRGS 838
Query: 311 -YNNRGRGGRGSYNNRGRGGDRSSVQCQICHKYGH 344
Y RGRG +N+RG +R +CQ+C GH
Sbjct: 839 NYRGRGRG----FNSRG---NRPPPRCQLCSLLGH 922
>BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element polyprotein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 608
Score = 55.5 bits (132), Expect = 2e-07
Identities = 31/135 (22%), Positives = 66/135 (47%), Gaps = 1/135 (0%)
Frame = +1
Query: 35 KLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRANSVENPA-FLVWEQHD 93
KL+ +N+ SW + + + F+ + P + V+ PA + +W + +
Sbjct: 172 KLNGTNYSSWSRSMVHALTAKNKVGFINGSIKTP----------SEVDQPAEYALWNRCN 321
Query: 94 SALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSITKGSKN 153
S + +WL S+ P + V++ + + Q+WE D F ++ Q++ L S+++G+ +
Sbjct: 322 SMILSWLTHSVEPDLAKGVIHAKTACQVWEDFKDQFSQKNIPAIYQIQKSLASLSQGTVS 501
Query: 154 TSDYLKRIKSIVNAL 168
S Y +IK + + L
Sbjct: 502 ASTYFTKIKGLWDEL 546
>BG582205
Length = 801
Score = 54.3 bits (129), Expect = 4e-07
Identities = 52/164 (31%), Positives = 78/164 (46%), Gaps = 15/164 (9%)
Frame = +3
Query: 652 LTTCKISVPHKSKYT-ICHSCCVAKSHRLPSSASSTLYTAPLELIFADLWGPASIESSAG 710
LT K+ + ++ K + C CC+ K+HRLPS + P + S
Sbjct: 342 LTVLKLHLVYQIKLSDFCSFCCLGKAHRLPS------------------FHPLYLTLSLL 467
Query: 711 YSYFLTCVDAFSR---------FTWIYLLKRKSETS--TVFIQFQAMVELKFGL--KIKS 757
S+F+ C D SR F++ + K+E S T+F F+ +VEL++ L ++ S
Sbjct: 468 NSFFVICGDYSSRVFFNLCGCLFSF*LDISFKTEVSHFTIFQNFKRIVELQYKLLHQVCS 647
Query: 758 VQTDGGTEFKPLLPHFLKLG-ITHRLTCPHTHHQNGSVERKHRH 800
GG EF+P + + LG H LT + GSVERKHRH
Sbjct: 648 NCLRGG-EFRPFITYLNNLG*PAHTLTT-----KMGSVERKHRH 761
>BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 514
Score = 52.4 bits (124), Expect = 1e-06
Identities = 30/80 (37%), Positives = 43/80 (53%), Gaps = 2/80 (2%)
Frame = -2
Query: 837 VLGNLSPYHKLFKVPADYKIL-RVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGY 895
VL + PY VP + R+FG + H+ SK R+ +CVF+ YSS+ KGY
Sbjct: 507 VLPSFYPY-----VPTSNNLTPRIFGCTSFVHIHSDGRSKFDHRALKCVFIRYSSTQKGY 343
Query: 896 KCLASDGRIY-ISKDVQFNE 914
+C R Y +S+DV F+E
Sbjct: 342 RCYHPPSRKYFVSRDVTFHE 283
>BQ154516
Length = 506
Score = 51.6 bits (122), Expect = 2e-06
Identities = 34/102 (33%), Positives = 54/102 (52%)
Frame = +1
Query: 567 QDTCKTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNN 626
+D+ + LL G +GSDGLY F K L +P S + SLS + ++++V S++
Sbjct: 232 KDSKQILLEGVVGSDGLYQF---------KPLQFLP-SMSKSLSYCNNATISSVVCNSSS 381
Query: 627 VPSPTAYDLWHSRLGHPHYETLQSALTTCKISVPHKSKYTIC 668
S +++ WH RLGH + ++S L CKI +K C
Sbjct: 382 -SSNDSFNKWHCRLGHANPNVVKSILNLCKIPFQNKHVLDFC 504
Score = 44.3 bits (103), Expect = 4e-04
Identities = 20/51 (39%), Positives = 29/51 (56%)
Frame = +3
Query: 455 PQTMLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQG 505
P AS+ + W+PDSGA+HH+T + ++ G +QV M NGQG
Sbjct: 84 PSEYAASSSYSEAPWYPDSGASHHLTFNPHNLAYRTPYNGQEQVLMDNGQG 236
>AW691018 weakly similar to GP|2462935|em open reading frame 1 {Brassica
oleracea}, partial (12%)
Length = 639
Score = 51.2 bits (121), Expect = 3e-06
Identities = 23/89 (25%), Positives = 48/89 (53%)
Frame = +2
Query: 77 RANSVENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQ 136
+A S ++P +W++ + + TW+L S+ P + +V+ + +W + D F ++
Sbjct: 299 KAPSADDPKLPLWKRCNDLVLTWILHSIEPDIARSVIFSDTAAAVWSDLHDRFSQGDESR 478
Query: 137 STQLRSELRSITKGSKNTSDYLKRIKSIV 165
Q+R E+ +GS SDY ++KS++
Sbjct: 479 IYQIRQEISECRQGSLLISDYYTKLKSLL 565
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 48.9 bits (115), Expect = 2e-05
Identities = 57/174 (32%), Positives = 77/174 (43%)
Frame = +2
Query: 1085 TIHGLWSLFPRAGIQ*GVNGFLESKRMQMGQ*IDIKPG*LPRVIIK*KALTMQKSFLQ** 1144
T HG+ LF +GF+E K+ QM ++KP * RV +K + K L **
Sbjct: 140 TKHGILFLFLHTRKLLAASGFIELKKTQMDLLTNLKPD*WLRVSVKHLDVITLKLSLL** 319
Query: 1145 SQSPFV*FSLWLFPKGRIFTNWM*IMLFFMGLSKKRFTCNNQLDFRILTKHLCAS*IKPY 1204
+ S FS L F + M F M K++ C N D ++L CAS*
Sbjct: 320 NLSLSGSFSPLLSLINGKFNKLISTMPF*MVFFKRKCICLNLRDLKLLINLWCAS*TNHS 499
Query: 1205 MGLNKLLGLGLTG*RQL*SSMGSKQVVVIPLSLLFTTSLMSSTY*SMWMILSSL 1258
M +K G+ * S+ S +VVVI L ++S * MWM SSL
Sbjct: 500 MA*SKHHVHGMRX*PVHKFSLVSLKVVVIHHYLSTIKMELASIX*YMWMTFSSL 661
>AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Danio rerio},
partial (14%)
Length = 510
Score = 46.6 bits (109), Expect = 8e-05
Identities = 54/217 (24%), Positives = 86/217 (38%), Gaps = 2/217 (0%)
Frame = +1
Query: 187 GLREDYSHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPP 246
GL E+Y+ + ++ R S+ + E++++ EA+ ++ R ++ S SA LAQ+ P
Sbjct: 1 GLLEEYNSFVLVIYIRLDSPSMEEGESILMMQEAQFEKYR-QELTNPSVSANLAQSELPS 177
Query: 247 QSTPQAPPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNR 306
+ P +SE +
Sbjct: 178 K--------------------------PGNSESQEVGTE--------------------- 216
Query: 307 DRGGYNNRGRG-GRGSYNNRGRGGDRSS-VQCQICHKYGHDASVCYYRGNSTSPATNSQP 364
Y N GRG GRG RGRG S+ +QCQIC + HDA+ C +R + A++SQ
Sbjct: 217 ----YYNAGRGRGRGRGRGRGRGRSNSNRLQCQICARNNHDAARCCFRYDQ---ASSSQA 375
Query: 365 NMTNQAPAPVAPTFGFGSSFGMMPQFGYSPFRGFQPY 401
+ ++AP S P G S F P+
Sbjct: 376 H--HRAPPSEYAASSSYSEAPWYPDSGASHHLTFNPH 480
Score = 33.9 bits (76), Expect = 0.51
Identities = 14/32 (43%), Positives = 19/32 (58%)
Frame = +1
Query: 449 ARHIRAPQTMLASAPSTLGNWHPDSGATHHVT 480
A H P AS+ + W+PDSGA+HH+T
Sbjct: 373 AHHRAPPSEYAASSSYSEAPWYPDSGASHHLT 468
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 43.1 bits (100), Expect = 8e-04
Identities = 34/125 (27%), Positives = 56/125 (44%)
Frame = +3
Query: 454 APQTMLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGS 513
A T PS W DSG THH+THD + ++ + +V M NG + ++ IG+
Sbjct: 69 AMSTFATKQPSKY--WLIDSGCTHHMTHDRDLFKE-LNKSTISKVRMLNGAHIEVEGIGT 239
Query: 514 ASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTL 573
S + + N+L P + ++L+SV + G F C + Q+ K +
Sbjct: 240 VLVKSHSGYKQ---ISNVLYAPKLNQSLLSVPQLL-TKGYKVLFEHEKCVIKDQNN-KEV 404
Query: 574 LRGSL 578
LR +
Sbjct: 405 LRAQM 419
>TC92013 homologue to GP|11595557|emb|CAC18142. related to c-module-binding
factor {Neurospora crassa}, partial (2%)
Length = 1437
Score = 42.7 bits (99), Expect = 0.001
Identities = 33/101 (32%), Positives = 40/101 (38%)
Frame = -2
Query: 231 AASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDR 290
AAS S + + P S P PP S+ PP P V + S++ S S +
Sbjct: 491 AASPSSVSSSDSGIPGASPPPPPPTSSSRRGRPPAGPTVVRLG--ESDLESESEISSSES 318
Query: 291 AYEDRDDDRSRSDDNRDRGGYNNRGRGGRGSYNNRGRGGDR 331
R R RGG RG GGRG RGRG R
Sbjct: 317 ESGIGGSPAMRGPSMRGRGGGRGRG-GGRGRGRGRGRGRGR 198
>BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l {Arabidopsis
thaliana}, partial (12%)
Length = 436
Score = 41.6 bits (96), Expect = 0.002
Identities = 30/105 (28%), Positives = 45/105 (42%)
Frame = -1
Query: 786 HTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYH 845
H H QNG E + I LL + P S W HA L A LI P+ SP
Sbjct: 388 HVHTQNGLAESFIKRIQLITRPLLMRSKQPVSAWGHAVLHAAELIRIRPSSE-HKYSPSQ 212
Query: 846 KLFKVPADYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSS 890
L D ++ FG Y + P + +K+ + + +++G+ S
Sbjct: 211 LLSGHVPDVSHIKTFGYPVYVPIAPPHRTKMGPQRRMGIYVGFDS 77
>AW773859
Length = 538
Score = 41.2 bits (95), Expect = 0.003
Identities = 31/105 (29%), Positives = 45/105 (42%), Gaps = 4/105 (3%)
Frame = -3
Query: 607 FSLSKPASPAVNTIVSTSNNVPSPTAYD---LWHSRLGHPHYETLQSALTTCK-ISVPHK 662
+ L+ +P+ S P PT LWH RLGH L S + I++
Sbjct: 323 YFLTTQDTPSATIASSQVQPQPQPTFLPQEALWHFRLGHLSNRKLLSLHSNFPFITIDQN 144
Query: 663 SKYTICHSCCVAKSHRLPSSASSTLYTAPLELIFADLWGPASIES 707
S ICH ++ +LP S+ + EL D+WGP S +S
Sbjct: 143 SVCDICH---YSRHKKLPFQLSTNRASKCYELFHFDIWGPFSTQS 18
>TC80054 SP|Q28009|FUS_BOVIN RNA-binding protein FUS (Pigpen protein).
[Bovine] {Bos taurus}, partial (4%)
Length = 988
Score = 40.8 bits (94), Expect = 0.004
Identities = 81/306 (26%), Positives = 119/306 (38%), Gaps = 18/306 (5%)
Frame = +1
Query: 241 QATAPPQSTPQAPPLVPSQSPAPPQAPPPVAIA-------PPSSEVNYISRTGSDDR--A 291
Q+TA Q + Q P P P P A PPS+ N + S D A
Sbjct: 7 QSTATTQQSTYDYYNQQPQQPQNPGGPAPPADGTAYNYSQPPSAGYNQPGQGYSQDGYGA 186
Query: 292 YEDRDDDRSRSDDNRDRG-GYNNRGRGGRGS-YNNRGRGGDRSSVQCQICHKYGHDASVC 349
Y+ + D+ GY + G + Y ++G+G D + V + G+
Sbjct: 187 YQQPPQSGYAQPTSYDQQQGYGSAQEGQTTTNYGSQGQG-DATQVPPVQPSQQGY----- 348
Query: 350 YYRGNSTSPATNSQPNMTNQAPAP---VAPTFGFGSSFGMMPQF--GYSPFRGFQPYGAP 404
G+S P+ N+ A P V PT +++G PQ GY P + +P G P
Sbjct: 349 ---GSSQQPSPNAAKYPPQGAAQPSYGVPPTSQ--TAYGSQPQTQSGYGPPQTQKPSGTP 513
Query: 405 SPFGMS-FPNSGFGSGSPYASGGMFTPPGYGFGSGYGFGLPPRAPARHIRAPQTMLASAP 463
+G S PN+ G G P P GY GY PP P+ + Q AP
Sbjct: 514 PAYGQSQSPNAAGGYGQPGYPPSQPPPSGYS-QPGYPPSQPP--PSGY---SQPGYGQAP 675
Query: 464 STLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSASFS-SPIHT 522
+ +SG+ A A S + +GN + G+ G P Q AS + SP
Sbjct: 676 QSY-----NSGSYGAGYPQAPAYSADGNASGNAR--GGSYDGAPAQGAQQASVAKSPQS* 834
Query: 523 QTTLLL 528
T++L+
Sbjct: 835 CTSMLI 852
>TC93151 homologue to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (4%)
Length = 657
Score = 40.8 bits (94), Expect = 0.004
Identities = 30/78 (38%), Positives = 39/78 (49%), Gaps = 2/78 (2%)
Frame = +3
Query: 353 GNSTSP-ATNSQPNMTNQAPAPVAPTFGFGSSFGMMPQFGYSPFRGFQPYG-APSPFGMS 410
G+S SP +T + P T AP+ +F FGSS + FG SP APS FG +
Sbjct: 198 GSSPSPFSTTTNPTSTATAPS----SFNFGSSSSL---FGASPASSTPAAASAPSIFGSA 356
Query: 411 FPNSGFGSGSPYASGGMF 428
S FG G+P G +F
Sbjct: 357 TAGSPFGGGTPSTPGNLF 410
Score = 32.3 bits (72), Expect = 1.5
Identities = 23/69 (33%), Positives = 30/69 (43%), Gaps = 1/69 (1%)
Frame = +3
Query: 353 GNSTSPATNSQPNMTNQAPAPVAPTFGFGSSFGMMPQFGYSPFRGFQP-YGAPSPFGMSF 411
G + S +T + M A + FGFGSS P FG P +GAPS S
Sbjct: 444 GGAASGSTTASTTMFGGASPATSTAFGFGSSASSTP*FGAPSSASSTPLFGAPS----ST 611
Query: 412 PNSGFGSGS 420
P+ G + S
Sbjct: 612 PSFGVAASS 638
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.331 0.142 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 59,163,148
Number of Sequences: 36976
Number of extensions: 1093399
Number of successful extensions: 15874
Number of sequences better than 10.0: 347
Number of HSP's better than 10.0 without gapping: 8123
Number of HSP's successfully gapped in prelim test: 427
Number of HSP's that attempted gapping in prelim test: 2025
Number of HSP's gapped (non-prelim): 12060
length of query: 1571
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1462
effective length of database: 4,984,343
effective search space: 7287109466
effective search space used: 7287109466
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0079b.1


