

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0074.19
(2281 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
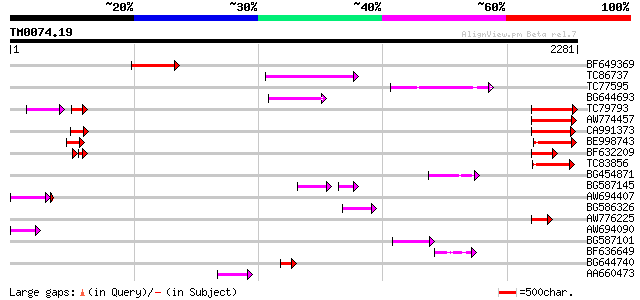
Score E
Sequences producing significant alignments: (bits) Value
BF649369 294 3e-79
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 253 6e-67
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 183 8e-46
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 168 3e-41
TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Ar... 155 1e-37
AW774457 weakly similar to GP|10176973|dbj gb|AAF19552.1~gene_id... 149 9e-36
CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imp... 145 1e-34
BE998743 similar to GP|22128591|gb| fertility restorer-like prot... 132 1e-30
BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01... 129 2e-29
TC83856 weakly similar to PIR|B96656|B96656 unknown protein 419... 127 4e-29
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 127 4e-29
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 108 2e-28
AW694407 weakly similar to GP|6630464|gb| F23N19.4 {Arabidopsis ... 113 4e-26
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 88 3e-17
AW776225 weakly similar to GP|18461169|dbj hypothetical protein~... 71 4e-12
AW694090 weakly similar to GP|11994279|dbj gb|AAD43616.1~gene_id... 70 1e-11
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 69 2e-11
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 68 4e-11
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 63 1e-09
AA660473 63 1e-09
>BF649369
Length = 631
Score = 294 bits (752), Expect = 3e-79
Identities = 150/195 (76%), Positives = 173/195 (87%)
Frame = +3
Query: 488 LQQQSLTLTELSKQMGRKETGHEGDTPIADSHPGESRLAGKKVKLPVFEGEDPVAWITRA 547
+QQQS+ + E++KQ+ ++T + + +S ESRLAGKKVKLP+FEG+DPVAWITRA
Sbjct: 3 MQQQSVLMAEVTKQLELQKTPLVREEYV-ESSQNESRLAGKKVKLPLFEGDDPVAWITRA 179
Query: 548 EIYFDVQNTPDEMRVKLTRLSMEGSTIHWFNLLMETEDNLSWEKLKRALIARYGGRRLEN 607
EIYFDVQNTPD+MRVKL+RLSMEG TIHWFNLLMETED+LS EKLK+ALIARY GRRLEN
Sbjct: 180 EIYFDVQNTPDDMRVKLSRLSMEGPTIHWFNLLMETEDDLSREKLKKALIARYDGRRLEN 359
Query: 608 PFEELSTLRQKGSVDEFVESFELLSSQVGRLPEDQYLGYFMSGLKPAIRRRVRTLNPGTR 667
PFEELSTLRQ GSV+EFVE+FELLSSQVGRLPE+QYLGYFMSGLK IRRRVRTLNP TR
Sbjct: 360 PFEELSTLRQIGSVEEFVEAFELLSSQVGRLPEEQYLGYFMSGLKAHIRRRVRTLNPTTR 539
Query: 668 MEMMRIAKDVEEELK 682
M+MMRIAKDVE+E +
Sbjct: 540 MQMMRIAKDVEDEFE 584
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 253 bits (646), Expect = 6e-67
Identities = 146/390 (37%), Positives = 222/390 (56%), Gaps = 17/390 (4%)
Frame = +1
Query: 1028 LEEFQEVFRSK--IQLPPERSKV-HQIKLFPEQETINVRP------YRYPHHQKEEIERQ 1078
LEEF ++F + Q+P R + H I L P+++ N P Y + +++
Sbjct: 346 LEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDG-NDPPLPWGPLYGMSRQELLVLKKT 522
Query: 1079 VAELMEAGIIRPSMSAYSSPVILVKKKDKSWRMCVDYRALNKATIPDKYPIPIVDELLDE 1138
+ +L++ G I+ S SA +PV+ V+K R CVDYRALN T D+YP+P++ E L
Sbjct: 523 LEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRR 702
Query: 1139 LNGASIFSKIDLKSGYHQIRVHEDDIEKTAFRTHNGHYEYLVMPFGLMNAPATFQAVMND 1198
+ GA F+K+D+ + +H++R+ ++D EKTAFRT G +E++V PFGL APATFQ +N
Sbjct: 703 VAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINK 882
Query: 1199 IFRPYLRKFVLVFFDDILIYSK-DLPEHLTHLKLVLSVLLANCFVANQTKCKFGCASIDY 1257
+L FV + DD+LIY+ +H ++ VL L + KC+F ++ Y
Sbjct: 883 TLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKY 1062
Query: 1258 LGHII-SGAGMAVDPEKVKCIMDWPVPKNVKGVRGFLGLTGYYRKFIKDYGKMANPLTEL 1316
+G I+ +G G++ DP K+ I DW P +VKG R FLG YY+ FI Y ++ PLT L
Sbjct: 1063 VGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRL 1242
Query: 1317 TKKD-SFSWGPEADRAFLQLKRVMTSPPVLILPNFTLPFEVECDAAGRGIGAVLMQQ--- 1372
T+KD F WG E + AF +LKR+ PVL + + VE D +G +G VL Q+
Sbjct: 1243 TRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDGT 1422
Query: 1373 --RQPLAFFSKALSAGNLAKSVYEKELMAL 1400
P+AF S+ LS +++KEL+A+
Sbjct: 1423 GAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 183 bits (464), Expect = 8e-46
Identities = 132/431 (30%), Positives = 205/431 (46%), Gaps = 18/431 (4%)
Frame = +2
Query: 1533 SPTSPSIPWLLEEFHGSPSGGHSGFLRTYRRLATTLYWVGMQKRVRDYVRACDVCQRHKY 1592
SP + L++E H S + GH G T ++ +W G + VR +VR CDVC
Sbjct: 152 SPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHI 331
Query: 1593 SALSPGGLLQPLPIPNAVWEDLSLDFITGLP--KSKGFEAVLVVVDRLSKYSHFILLKHP 1650
+ G L+PLP+PN + DLS+DFIT LP + +G + + V+VDRLSK + L+
Sbjct: 332 WRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKS---VTLEEM 502
Query: 1651 YT--AKSIAEVFVREVVRLHGIPNSVISDRDPIFVSHFWSELFKLQGTKLKMSSAYHPET 1708
T A++ A+ F+ R HG+P S++SDR +V FW E +L G +S++YHP+T
Sbjct: 503 DTMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQT 682
Query: 1709 DGQTEVINRCLESYLRCFASDHPKSWSHWISWAEFWYNTTFHSSIGQTPFEVVYGRQAPP 1768
DG TE N+ +++ LR + +W + + +SSIG TPF V +G P
Sbjct: 683 DGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYHVDP 862
Query: 1769 IVKFLSNETKV----AAVALELSERDEALRQLRGHLQKAQEQMAIYANKKRRDLS-FAVG 1823
I V AA L + + ++ + AQ++ ANK+R + VG
Sbjct: 863 IPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEASANKRRCPADRYQVG 1042
Query: 1824 EWVFLKLRPHRQHSVVKRI----NQKLAARFYGPFQIEAKVGAVAYRLKLPAESKIHPVF 1879
+ V+L + ++ K++ ++ RF P +E V Y P F
Sbjct: 1043 DKVWLNVSNYKSPRPSKKLDWLHHKYEVTRFVTPHVVELNVPGTVY-----------PKF 1189
Query: 1880 HISLLKKAV-----GNYQVQGQLPADLGVDDTTDIYPEVIVGTRTVRLGDSEVHQSLVKW 1934
H+ LL++A G V Q P + D + E I+ R ++G Q+LVKW
Sbjct: 1190 HVDLLRRAASDPLPGQEVVDPQPPPIVDDDGEVEWEVEEILAARWHQVGRGRRRQALVKW 1369
Query: 1935 KHKSMEEVTWE 1945
K + TWE
Sbjct: 1370 --KGFVDATWE 1396
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 168 bits (425), Expect = 3e-41
Identities = 99/236 (41%), Positives = 136/236 (56%), Gaps = 3/236 (1%)
Frame = +2
Query: 1041 LPPERSKVHQIKLFPEQETINVRPYRYPHHQKEEIERQVAELMEAGIIRPSMSAYSSPVI 1100
+PPE I L P I + YR + + ++ Q+ +L+E G I+PS+ V+
Sbjct: 17 VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVL 196
Query: 1101 LVKKKDKSWRMCVDYRALNKATIPDKYPIPIVDELLDELNGASIFSKIDLKSGYHQIRVH 1160
+KKKD RM +DY LN I KYP+P++DEL D L G+ F KIDL+ G HQ RV
Sbjct: 197 FLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVI 376
Query: 1161 EDDIEKTAFRTHNGHYEYLVMPFGLMNAPATFQAVMNDIFRPYLRKFVLVFFDDILIYSK 1220
+D+ KTAFR GHYE LVM FG N P F +MN +F+ YL V+VF +DILIYSK
Sbjct: 377 GEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSK 556
Query: 1221 DLPEHLTHLKLVLSVL--LANCFVANQ-TKCKFGCASIDYLGHIISGAGMAVDPEK 1273
+ EH HL+L L VL + C ++ + G S+ H+ISG G+ VD ++
Sbjct: 557 NENEHENHLRLALKVLKDIGLCQISYV*ILVEVGFFSL----HVISGEGLKVDSKR 712
>TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Arabidopsis
thaliana}, partial (7%)
Length = 1580
Score = 155 bits (393), Expect = 1e-37
Identities = 102/185 (55%), Positives = 116/185 (62%)
Frame = +3
Query: 2097 QNGLQKGCS*YSYLQFSY*WFMQNRESLSCLGAS**DACEGSTSQYNHL*FFIGCFMQEP 2156
+N QK S* LQF Y*WF Q RE+L C A DA T+ Y +L F+I CFMQ P
Sbjct: 735 RNASQKHYS*CRNLQFPY*WFKQIRENLICFAACRSDA**RCTT*YMYLQFYIRCFMQNP 914
Query: 2157 SC*QSNCIDQEN*RPGCSARFMHIQYTYGWTMQSWKT*GCARDFS*SFDKRLSYNCPYIY 2216
S *QSNCI E *R G S R++HIQY+Y T+ WKT GC F SF +R C YIY
Sbjct: 915 SS*QSNCIVNEI*RQGFSTRYIHIQYSYQRTVPKWKTGGCT*SF*RSFGQRP*SKCRYIY 1094
Query: 2217 CYDQWTL*RRLV**SVGLAVKNGN*GLYS*CYNF*IYYPFTL*KW*E*QG*ETSS*ND** 2276
YD L R LV**SVGLAVKNG L+S C N * YPF++ K *+* G ETSS*ND
Sbjct: 1095 HYDPRVLCRGLV**SVGLAVKNGRQWLHSRC*NL*NNYPFSVQKR*K*YGRETSS*NDRE 1274
Query: 2277 RSTGK 2281
RST K
Sbjct: 1275 RSTVK 1289
Score = 101 bits (252), Expect(2) = 3e-35
Identities = 69/151 (45%), Positives = 85/151 (55%)
Frame = +3
Query: 68 FSREVISQTPSPSLLSSKVSVFTARFKEHYNFMMTC*LRDFY*TKSVTGP*SMGYVNWGK 127
FS+ V Q P PSL S++V V R +H++FM+ DF TK GP*SM YV K
Sbjct: 3 FSKWVTCQIP*PSLHSARVFVSKVRSSKHFSFMIRWWRLDFISTKLAMGP*SMAYVR*EK 182
Query: 128 QVLLWSCLGGFKGEGLNLMW*CTTPSSIACVKISL*VMLLIYILKWLPREYLLM*SRTVL 187
Q + +LMW*CTTP IAC K++L + LLIY LKW R +L M TVL
Sbjct: 183 QEQPLTYSNELMVIWFSLMW*CTTPLLIACAKLNLLMKLLIYFLKWFLRGFLPMLLLTVL 362
Query: 188 *YMAFALLVN*NKQLGC*MKWD*KT*TQMFI 218
* + FA LVN* QL C +K KT +M I
Sbjct: 363 *LVVFAFLVN*KMQLICLIK*YWKTSNRMCI 455
Score = 67.8 bits (164), Expect(2) = 3e-35
Identities = 40/65 (61%), Positives = 49/65 (74%)
Frame = +2
Query: 249 NLMLLLIIP*WMGIA*LMK*IKPKMYSMLCLNEERLQMFIATIS*STDYVRLKWLMKP*I 308
NL+LLLI *WM IA*L K*I+ ++YS L L E + +F T+S* T +VRLK LMKP*I
Sbjct: 548 NLILLLITL*WMDIA*LKK*IRLRVYSTLWLRGE*ILIFRVTVS*LTGFVRLKSLMKP*I 727
Query: 309 SLQKC 313
SL+KC
Sbjct: 728 SLKKC 742
>AW774457 weakly similar to GP|10176973|dbj
gb|AAF19552.1~gene_id:MTG13.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (13%)
Length = 650
Score = 149 bits (377), Expect = 9e-36
Identities = 100/179 (55%), Positives = 117/179 (64%)
Frame = -3
Query: 2100 LQKGCS*YSYLQFSY*WFMQNRESLSCLGAS**DACEGSTSQYNHL*FFIGCFMQEPSC* 2159
+Q+ CS* L FS+*WF+Q RE+L CLGA **+A STSQY HL F CFM +PSC*
Sbjct: 585 VQRNCS**GNL*FSH*WFVQIRENLLCLGAC**NA**WSTSQYIHLQLFNRCFM*KPSC* 406
Query: 2160 QSNCIDQEN*RPGCSARFMHIQYTYGWTMQSWKT*GCARDFS*SFDKRLSYNCPYIYCYD 2219
CI EN*R S R++H+QYTY W MQS T* C R F SF+ L C + Y
Sbjct: 405 PGYCIS*EN*RSRHSTRYVHVQYTYLWIMQSRTT*ECTRCFPGSFE*GL*CKCLDV*YYG 226
Query: 2220 QWTL*RRLV**SVGLAVKNGN*GLYS*CYNF*IYYPFTL*KW*E*QG*ETSS*ND**RS 2278
QWTL*R LV**S GL VKNG +S*C N * +P +L *E*+G ETS+*ND *RS
Sbjct: 225 QWTL*RGLV**SRGLVVKNG*QWYHS*CCNL*NSHPGSLS*R*E*KGGETSA*NDC*RS 49
Score = 34.3 bits (77), Expect = 0.57
Identities = 14/18 (77%), Positives = 16/18 (88%)
Frame = -2
Query: 296 DYVRLKWLMKP*ISLQKC 313
D+VRLKWLMKP*+SL C
Sbjct: 640 DFVRLKWLMKP*VSLMIC 587
>CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 756
Score = 145 bits (367), Expect = 1e-34
Identities = 92/178 (51%), Positives = 114/178 (63%)
Frame = +1
Query: 2097 QNGLQKGCS*YSYLQFSY*WFMQNRESLSCLGAS**DACEGSTSQYNHL*FFIGCFMQEP 2156
+N Q S* LQF Y*WF+ E+L C AS**DA G T+ YN+L F+IGCFMQ+P
Sbjct: 205 RNASQTYYS*CGNLQFPY*WFV*IGENLLCFEAS**DA*SGCTT*YNYLQFYIGCFMQKP 384
Query: 2157 SC*QSNCIDQEN*RPGCSARFMHIQYTYGWTMQSWKT*GCARDFS*SFDKRLSYNCPYIY 2216
S *Q NC+ + RPG S +++HI Y+Y WT+ WKT C F SF +RL YNC +IY
Sbjct: 385 SG*QGNCVINKIERPGYSTKYVHIHYSY*WTV*RWKTRRCT*YF*GSFGQRL*YNCQHIY 564
Query: 2217 CYDQWTL*RRLV**SVGLAVKNGN*GLYS*CYNF*IYYPFTL*KW*E*QG*ETSS*ND 2274
YD W L*+ LV**S+ VKN L+S C N * +PF +* *+* G ET S*ND
Sbjct: 565 RYDPWVL*QGLV**SIDFVVKNERQ*LHSRCCNL*NNHPFPV**R*K**GGET-S*ND 735
Score = 63.5 bits (153), Expect = 9e-10
Identities = 40/73 (54%), Positives = 49/73 (66%)
Frame = +3
Query: 245 NKV*NLMLLLIIP*WMGIA*LMK*IKPKMYSMLCLNEERLQMFIATIS*STDYVRLKWLM 304
NK NL+LL I *WM IA*L K*I+ ++Y LCL E + +F TIS* +VRLK M
Sbjct: 6 NKALNLLLLPIAL*WMDIA*LKK*IRLRVYYTLCLRGE*ILIFKVTIS*LMGFVRLKRSM 185
Query: 305 KP*ISLQKCDRNL 317
K *ISL+KC N+
Sbjct: 186 KL*ISLKKCITNI 224
Score = 32.0 bits (71), Expect = 2.8
Identities = 29/89 (32%), Positives = 43/89 (47%)
Frame = +3
Query: 114 VTGP*SMGYVNWGKQVLLWSCLGGFKGEGLNLMW*CTTPSSIACVKISL*VMLLIYILKW 173
VT * MG+V + + L L L LMW* T P + CV +ML +++
Sbjct: 138 VTIS*LMGFVRLKRSMKL*ISLKKCITNILFLMW*LTIPLLMVCVNWGKSLML*S*LMRC 317
Query: 174 LPREYLLM*SRTVL*YMAFALLVN*NKQL 202
+ Y L+* TVL +M +A + +QL
Sbjct: 318 MIGVYHLI*LLTVLYWMLYAKTIRLTRQL 404
>BE998743 similar to GP|22128591|gb| fertility restorer-like protein {Petunia x
hybrida}, partial (7%)
Length = 829
Score = 132 bits (333), Expect = 1e-30
Identities = 94/174 (54%), Positives = 109/174 (62%)
Frame = +1
Query: 2105 S*YSYLQFSY*WFMQNRESLSCLGAS**DACEGSTSQYNHL*FFIGCFMQEPSC*QSNCI 2164
S* S L + *W + N L CL A *DAC ST+ + +L FF CFMQ+ S *QS CI
Sbjct: 274 S*CSEL*YHG*WIL*N---LPCLEACR*DAC*WSTT*HIYLQFFNRCFMQKQSS*QSYCI 444
Query: 2165 DQEN*RPGCSARFMHIQYTYGWTMQSWKT*GCARDFS*SFDKRLSYNCPYIYCYDQWTL* 2224
QEN*RP S +++HIQYTY W MQ T* C R FS S + L P + +DQWTL*
Sbjct: 445 SQEN*RPRHSTKYVHIQYTY*WVMQRRTT*ECTRCFSRSIN*GL*SKHPNV*YFDQWTL* 624
Query: 2225 RRLV**SVGLAVKNGN*GLYS*CYNF*IYYPFTL*KW*E*QG*ETSS*ND**RS 2278
R LV**S GL VKNG * S*C N * YYP +L +*+G ETSS*N *RS
Sbjct: 625 RGLV**SRGLVVKNGG**YQS*CCNL*NYYPLSLL*RLQ*KGGETSS*NGC*RS 786
Score = 50.1 bits (118), Expect = 1e-05
Identities = 38/70 (54%), Positives = 44/70 (62%)
Frame = +3
Query: 229 RKGR*EKLRTC*L**LNKV*NLMLLLIIP*WMGIA*LMK*IKPKMYSMLCLNEERLQMFI 288
+K *++LR L** +V*N MLL I *WMGIA*LMK*I+P M+ L L E L M
Sbjct: 108 KKEM*KELRMRWL**SKEV*NQMLLHTIH*WMGIA*LMK*IRPNMF*ALLLEWEWLLMLR 287
Query: 289 ATIS*STDYV 298
A IS D V
Sbjct: 288 AIISWLMDSV 317
>BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01051 protein
domain and multiple PPR PF|01535 repeats. EST
gb|AA728420 comes, partial (13%)
Length = 508
Score = 129 bits (323), Expect = 2e-29
Identities = 69/104 (66%), Positives = 75/104 (71%)
Frame = +3
Query: 2098 NGLQKGCS*YSYLQFSY*WFMQNRESLSCLGAS**DACEGSTSQYNHL*FFIGCFMQEPS 2157
N LQK CS*Y LQFS *WF+Q E+L CLGA **DA STS YNHL F CFMQ+PS
Sbjct: 192 NALQKNCS*YGNLQFSN*WFVQIWENLLCLGAC**DA**WSTS*YNHLQFINRCFMQKPS 371
Query: 2158 C*QSNCIDQEN*RPGCSARFMHIQYTYGWTMQSWKT*GCARDFS 2201
C*Q CI QEN*RP S R++HIQYTY WTMQ+ K * C DFS
Sbjct: 372 C*QGYCISQEN*RPRHSTRYVHIQYTYRWTMQTRKA*RCTSDFS 503
Score = 40.0 bits (92), Expect(2) = 1e-04
Identities = 21/39 (53%), Positives = 25/39 (63%)
Frame = +2
Query: 275 SMLCLNEERLQMFIATIS*STDYVRLKWLMKP*ISLQKC 313
S+L L E L M +A +S* D VRLKW KP*+S KC
Sbjct: 80 SILYLE*EWLPMLVAIVS*LMDSVRLKWSTKP*VSFMKC 196
Score = 25.4 bits (54), Expect(2) = 1e-04
Identities = 13/23 (56%), Positives = 15/23 (64%)
Frame = +3
Query: 251 MLLLIIP*WMGIA*LMK*IKPKM 273
MLL + *WM I LMK I+P M
Sbjct: 6 MLLPTVH*WMDIVLLMKLIRPNM 74
>TC83856 weakly similar to PIR|B96656|B96656 unknown protein 41955-40111
[imported] - Arabidopsis thaliana, partial (12%)
Length = 662
Score = 127 bits (320), Expect = 4e-29
Identities = 83/171 (48%), Positives = 105/171 (60%)
Frame = +1
Query: 2101 QKGCS*YSYLQFSY*WFMQNRESLSCLGAS**DACEGSTSQYNHL*FFIGCFMQEPSC*Q 2160
Q GC + Y+ * F+ E+L C AS *DA ST+ Y++L F+IGCFMQ PS *Q
Sbjct: 70 QTGCVYF*YIG---*CFL*IGENLLCFEASG*DA**RSTT*YSYLQFYIGCFMQNPSG*Q 240
Query: 2161 SNCIDQEN*RPGCSARFMHIQYTYGWTMQSWKT*GCARDFS*SFDKRLSYNCPYIYCYDQ 2220
+CI + RP S ++ HI Y+Y W + WKT GC F SF + L YNC +IY Y
Sbjct: 241 GSCIINKIERPRYSTKYAHIHYSY*WIVYKWKTGGCTEYF*GSFSQGL*YNCRHIYRYVL 420
Query: 2221 WTL*RRLV**SVGLAVKNGN*GLYS*CYNF*IYYPFTL*KW*E*QG*ETSS 2271
W L* LV**S+GLAVKNG ++S C N *I F++ K *+* G ETSS
Sbjct: 421 WVL*EGLV**SIGLAVKNGREWMHSRC*NL*IN*AFSVQKR*K*YGRETSS 573
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 127 bits (320), Expect = 4e-29
Identities = 73/207 (35%), Positives = 113/207 (54%)
Frame = +2
Query: 1684 SHFWSELFKLQGTKLKMSSAYHPETDGQTEVINRCLESYLRCFASDHPKSWSHWISWAEF 1743
S+FW +LFKL GT L MSSAYHP +DGQ+E +N+ E YLRC P WS WAE+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1744 WYNTTFHSSIGQTPFEVVYGRQAPPIVKFLSNETKVAAVALELSERDEALRQLRGHLQKA 1803
WYNT+++ S TPF+ +YGR +++ + A + +L++R+E L QL+ +
Sbjct: 212 WYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQREELLSQLQSISTRL 391
Query: 1804 QEQMAIYANKKRRDLSFAVGEWVFLKLRPHRQHSVVKRINQKLAARFYGPFQIEAKVGAV 1863
+ +I K R+ + WV L + ++ + + + P + A
Sbjct: 392 NKL*SI---KLIRNAAILSSSWVSTSL*NCNLINSLR*HCGNIKSSVH-PTLVHY*QSA- 556
Query: 1864 AYRLKLPAESKIHPVFHISLLKKAVGN 1890
AY+L LP+ +K+ P+FH+S LK GN
Sbjct: 557 AYKLSLPSTAKVPPIFHVSQLKPFHGN 637
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 108 bits (271), Expect(2) = 2e-28
Identities = 56/136 (41%), Positives = 81/136 (59%)
Frame = +2
Query: 1159 VHEDDIEKTAFRTHNGHYEYLVMPFGLMNAPATFQAVMNDIFRPYLRKFVLVFFDDILIY 1218
+H DD+EKTAF T G Y Y VMPFGL NA +T+Q ++N +F L + V+ DD+L+
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 1219 SKDLPEHLTHLKLVLSVLLANCFVANQTKCKFGCASIDYLGHIISGAGMAVDPEKVKCIM 1278
S +HL HLK L N KC FG S ++LG+I++ G+ V+P+++ I+
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 1279 DWPVPKNVKGVRGFLG 1294
D P PKN + V+ G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 37.7 bits (86), Expect(2) = 2e-28
Identities = 22/86 (25%), Positives = 42/86 (48%), Gaps = 4/86 (4%)
Frame = +3
Query: 1322 FSWGPEADRAFLQLKRVMTSPPVLILPNFTLPFEVECDAAGRGIGAVLMQ----QRQPLA 1377
F W + + AF QLK+ +T+PPVL P + + + +VL++ +++P+
Sbjct: 495 FVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIAISSTAVSSVLIREDRGEQKPIF 674
Query: 1378 FFSKALSAGNLAKSVYEKELMALVLS 1403
+ SK ++ EK A++ S
Sbjct: 675 YTSKRMTDPETRYPTLEKMAFAVITS 752
>AW694407 weakly similar to GP|6630464|gb| F23N19.4 {Arabidopsis thaliana},
partial (4%)
Length = 661
Score = 113 bits (282), Expect(2) = 4e-26
Identities = 76/161 (47%), Positives = 92/161 (56%)
Frame = +2
Query: 2 LLPHPSSNSTRF*LHLLRRSITPPPFHFLTKWSSPESSQALLL*TS*SIATAT*VKSIPL 61
+LPHP N TRF L L + SITP FHF+ KW+ E SQ L TS SIA V ++ L
Sbjct: 38 ILPHPPFNLTRFYLPL*KLSITPLLFHFINKWN*MELSQISSLSTSSSIAFLNWV*TLFL 217
Query: 62 FLYSPEFSREVISQTPSPSLLSSKVSVFTARFKEHYNFMMTC*LRDFY*TKSVTGP*SMG 121
FLYSP FS+ VI+Q P S+ SS+VSV +H FM+ * RDF TK GP*
Sbjct: 218 FLYSPRFSKRVITQLP*HSIHSSRVSVSKVTSIKHCTFMIRW*HRDFIWTKLAMGP*LTD 397
Query: 122 YVNWGKQVLLWSCLGGFKGEGLNLMW*CTTPSSIACVKISL 162
YV + LL +C G +LM *CTT I C K+SL
Sbjct: 398 YVKLDE*QLLSNC*NELMGN*FSLMR*CTT*LLIICAKLSL 520
Score = 25.4 bits (54), Expect(2) = 4e-26
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = +3
Query: 163 *VMLLIYILKWL 174
*+MLLIYILKWL
Sbjct: 522 *LMLLIYILKWL 557
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 88.2 bits (217), Expect = 3e-17
Identities = 48/134 (35%), Positives = 75/134 (55%)
Frame = +2
Query: 1340 TSPPVLILPNFTLPFEVECDAAGRGIGAVLMQQRQPLAFFSKALSAGNLAKSVYEKELMA 1399
TS P+L+LP + + V DA+ G+G VL Q + +A+ S+ L ++ E+ A
Sbjct: 8 TSAPILVLPEL-ITYVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 1400 LVLSIQHWRHYLLGKEFIVYTDHKSLKHFLQQKVSSPDQQCWLAKLMGYQFQVKYKPGLE 1459
+V +++ WR YL G + ++TDHKSLK+ Q + Q+ W+ + Y + Y PG
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 1460 NKAADALSRRYDEV 1473
N ADALSRR +V
Sbjct: 365 NLVADALSRRRVDV 406
>AW776225 weakly similar to GP|18461169|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 5 MCL19.15, partial
(10%)
Length = 396
Score = 71.2 bits (173), Expect = 4e-12
Identities = 38/87 (43%), Positives = 53/87 (60%)
Frame = +3
Query: 2097 QNGLQKGCS*YSYLQFSY*WFMQNRESLSCLGAS**DACEGSTSQYNHL*FFIGCFMQEP 2156
+N Q S* L F Y*W +Q +++ C G DA +T+ YNH+ F+IGC MQ+P
Sbjct: 135 RNASQTNFS*CDNL*FPY*WIVQIWKNIICFGTYRFDA*SRATT*YNHVQFYIGCLMQKP 314
Query: 2157 SC*QSNCIDQEN*RPGCSARFMHIQYT 2183
S *Q NCI + RPG S +++HI Y+
Sbjct: 315 SG*QGNCIINKIERPGYSTKYVHIHYS 395
Score = 43.1 bits (100), Expect = 0.001
Identities = 24/46 (52%), Positives = 30/46 (65%)
Frame = +2
Query: 271 PKMYSMLCLNEERLQMFIATIS*STDYVRLKWLMKP*ISLQKCDRN 316
P++Y LCL E L +F ATI * D+VRLKW +KP* +KC N
Sbjct: 14 PRVY*TLCLIGE*LLLFGATIL*LMDFVRLKWWIKP*NFSKKCITN 151
>AW694090 weakly similar to GP|11994279|dbj
gb|AAD43616.1~gene_id:F16J14.3~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (4%)
Length = 659
Score = 69.7 bits (169), Expect = 1e-11
Identities = 52/122 (42%), Positives = 63/122 (51%)
Frame = +3
Query: 2 LLPHPSSNSTRF*LHLLRRSITPPPFHFLTKWSSPESSQALLL*TS*SIATAT*VKSIPL 61
+L H SSNST+F L L T FHF+ KW+ E Q L S SI + V S+ L
Sbjct: 186 ILHHTSSNSTKFYLPLSSSINTLLLFHFIHKWN*TELLQTWSLAASSSILSVNWVTSLLL 365
Query: 62 FLYSPEFSREVISQTPSPSLLSSKVSVFTARFKEHYNFMMTC*LRDFY*TKSVTGP*SMG 121
FL+ FS+ V SQ P PSL SS+V V +H FM+ DF TK V P* M
Sbjct: 366 FLFWQRFSKTVTSQIP*PSLHSSRVFVSKVTSIKHCTFMIRLLRWDFIWTKLVMEP*LMA 545
Query: 122 YV 123
V
Sbjct: 546 CV 551
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 68.9 bits (167), Expect = 2e-11
Identities = 47/170 (27%), Positives = 79/170 (45%), Gaps = 1/170 (0%)
Frame = +2
Query: 1539 IPWLLEEFHGSPSGGHSGFLRTYRRLATT-LYWVGMQKRVRDYVRACDVCQRHKYSALSP 1597
IP +L HGS GH +T ++ +W M K ++ CD CQR +
Sbjct: 95 IPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*R- 271
Query: 1598 GGLLQPLPIPNAVWEDLSLDFITGLPKSKGFEAVLVVVDRLSKYSHFILLKHPYTAKSIA 1657
+ Q + V++ +DF+ P S + +LV VD +SK+ I A +
Sbjct: 272 NEMPQNFILEVEVFDVWGIDFMGPFPSSYNNKYILVAVDYVSKWVEAIA-SPTNDATVVV 448
Query: 1658 EVFVREVVRLHGIPNSVISDRDPIFVSHFWSELFKLQGTKLKMSSAYHPE 1707
++F + G+P VISD F++ + +L K G + K+++AYHP+
Sbjct: 449 KMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 68.2 bits (165), Expect = 4e-11
Identities = 54/179 (30%), Positives = 87/179 (48%), Gaps = 9/179 (5%)
Frame = +1
Query: 1707 ETDGQTEVINRCLESYLRCFASDHPKSWSHWISWAEFWYNTTFHSSIGQTPFEVVYGRQA 1766
++D Q ++N LE++L F S+ + +++WAE YNT FH + G TPF+VVY
Sbjct: 4 DSDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY---- 171
Query: 1767 PPIVKFLSNETKVAAVALELSERDEALRQLR-----GHLQKAQEQMAIYANK----KRRD 1817
+V L VA +L R+E L + G +A E ++ A + RR
Sbjct: 172 --VVAHLQK----FVVARDLIYRNEGLHKSST*TSFGRGTRAYEALSRPAYETC*HPRRP 333
Query: 1818 LSFAVGEWVFLKLRPHRQHSVVKRINQKLAARFYGPFQIEAKVGAVAYRLKLPAESKIH 1876
LS R + ++ K A YGP+Q+ ++G+VA++L LP + +IH
Sbjct: 334 LSIVY----------TRDRTYEWQVLPKYVA*CYGPYQVIKQIGSVAFKL*LPEQHQIH 480
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 62.8 bits (151), Expect = 1e-09
Identities = 28/65 (43%), Positives = 42/65 (64%)
Frame = -1
Query: 1089 RPSMSAYSSPVILVKKKDKSWRMCVDYRALNKATIPDKYPIPIVDELLDELNGASIFSKI 1148
+PS+S + ++ V+KKD +RMC+DYR NK T +KYP+P +D L D++ F I
Sbjct: 262 QPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NI 83
Query: 1149 DLKSG 1153
DL+ G
Sbjct: 82 DLRLG 68
>AA660473
Length = 655
Score = 62.8 bits (151), Expect = 1e-09
Identities = 48/151 (31%), Positives = 73/151 (47%), Gaps = 9/151 (5%)
Frame = -2
Query: 836 EDEAVVEYKLMGVLGRMEEHH-------TMKIEGQVDNVNLLVLIDSGASHNFISPAVTN 888
E E ++ Y MG+ E + T+++ G + V +++L+D GA+HNF + +
Sbjct: 627 EKEDILVYNSMGLCEMTEFNKLKVVKPATLQLVGCLKGVPIVILVDIGANHNFDFASSVS 448
Query: 889 ALGLVITPIASRHIRLGDGHRVVTQGVCKGIKARLGKIEV-VIDALVLELGGLDMVLGVS 947
+G I S+ +G + G + + +DA LELG DM LGV+
Sbjct: 447 DVGCKGRVILSQESEVG*WSENINVG*AY*SRGTVRGFHC*GVDAYELELGEFDMFLGVA 268
Query: 948 WLSTLGK-VVMDWKLLTMQFVHGNQVVKLQG 977
WL LG VV DW T+ F +VVKLQG
Sbjct: 267 WLEKLGN*VVFDWDERTICFEWKGEVVKLQG 175
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.336 0.147 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,804,353
Number of Sequences: 36976
Number of extensions: 1038130
Number of successful extensions: 8136
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 4327
Number of HSP's successfully gapped in prelim test: 390
Number of HSP's that attempted gapping in prelim test: 3412
Number of HSP's gapped (non-prelim): 5290
length of query: 2281
length of database: 9,014,727
effective HSP length: 112
effective length of query: 2169
effective length of database: 4,873,415
effective search space: 10570437135
effective search space used: 10570437135
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0074.19


