

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
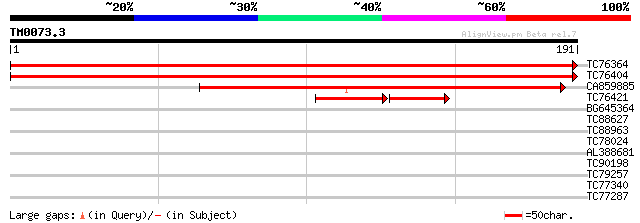
Query= TM0073.3
(191 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76364 similar to GP|4588906|gb|AAD26256.1| ribosomal protein S... 350 1e-97
TC76404 similar to GP|4588906|gb|AAD26256.1| ribosomal protein S... 350 2e-97
CA859885 weakly similar to EGAD|80130|866 probable ribosomal pro... 141 2e-34
TC76421 homologue to PIR|T02995|T02995 unspecific monooxygenase ... 50 5e-13
BG645364 32 0.15
TC88627 apyrase-like protein [Medicago truncatula] 32 0.20
TC88963 similar to GP|21554778|gb|AAM63688.1 unknown {Arabidopsi... 30 0.44
TC78024 similar to GP|20804963|dbj|BAB92640. hypothetical protei... 30 0.57
AL388681 similar to PIR|S12789|S127 GTP-binding protein ryh1 - f... 29 0.98
TC90198 similar to GP|4468835|emb|CAB38221.1 hairless {Drosophil... 27 3.7
TC79257 weakly similar to GP|18086351|gb|AAL57638.1 AT5g12890/T2... 27 6.3
TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog ... 27 6.3
TC77287 SP|Q02735|CAPP_MEDSA Phosphoenolpyruvate carboxylase (EC... 26 8.3
>TC76364 similar to GP|4588906|gb|AAD26256.1| ribosomal protein S7 {Secale
cereale}, partial (98%)
Length = 834
Score = 350 bits (899), Expect = 1e-97
Identities = 176/191 (92%), Positives = 186/191 (97%)
Frame = +1
Query: 1 MYTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKA 60
M+TSRKKI KDK AEPTEFEE+VGQALFDLENT+ ELKS+LKDLYINSAVQVDVSGNRKA
Sbjct: 67 MFTSRKKISKDKGAEPTEFEESVGQALFDLENTNHELKSELKDLYINSAVQVDVSGNRKA 246
Query: 61 VVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLT 120
VVIHVPYRLRK FRK+HV+LVRELEKKFSGKDVILIATRRI+RPPKKGSAAQRPR+RTLT
Sbjct: 247 VVIHVPYRLRKGFRKIHVRLVRELEKKFSGKDVILIATRRIVRPPKKGSAAQRPRSRTLT 426
Query: 121 AVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 180
AVHEAMLEDVVLPAEIVGKR RYRVDGSKIMKVFLDPKERNNTEYKLETF+AVYRKLSGK
Sbjct: 427 AVHEAMLEDVVLPAEIVGKRTRYRVDGSKIMKVFLDPKERNNTEYKLETFSAVYRKLSGK 606
Query: 181 DVVFEYPITEA 191
DVVFEYP+TEA
Sbjct: 607 DVVFEYPVTEA 639
>TC76404 similar to GP|4588906|gb|AAD26256.1| ribosomal protein S7 {Secale
cereale}, partial (98%)
Length = 909
Score = 350 bits (898), Expect = 2e-97
Identities = 175/191 (91%), Positives = 186/191 (96%)
Frame = +3
Query: 1 MYTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKA 60
M+TSRKKI KDK AEPTEFEE+VGQALFDLENT+ ELKS+LKDLYINSAVQVDVSGNRKA
Sbjct: 123 MFTSRKKISKDKGAEPTEFEESVGQALFDLENTNHELKSELKDLYINSAVQVDVSGNRKA 302
Query: 61 VVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLT 120
VVIHVPYRLRK FRK+HV+LVRELEKKFSGKDVILIATRRI+RPPKKGSAAQRPR+RTLT
Sbjct: 303 VVIHVPYRLRKGFRKIHVRLVRELEKKFSGKDVILIATRRIVRPPKKGSAAQRPRSRTLT 482
Query: 121 AVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 180
AVHEAMLEDVVLPAEIVGKR RYR+DGSKIMKVFLDPKERNNTEYKLETF+AVYRKLSGK
Sbjct: 483 AVHEAMLEDVVLPAEIVGKRTRYRIDGSKIMKVFLDPKERNNTEYKLETFSAVYRKLSGK 662
Query: 181 DVVFEYPITEA 191
DVVFEYP+TEA
Sbjct: 663 DVVFEYPVTEA 695
>CA859885 weakly similar to EGAD|80130|866 probable ribosomal protein S7
{Schizosaccharomyces pombe}, partial (60%)
Length = 522
Score = 141 bits (355), Expect = 2e-34
Identities = 72/125 (57%), Positives = 91/125 (72%), Gaps = 2/125 (1%)
Frame = +1
Query: 65 VPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQ--RPRTRTLTAV 122
VP L K F KV +L RELEKKFS + ++ IA RRILR P + S + RPR+RTLT+V
Sbjct: 1 VPVPLLKQFHKVQQRLTRELEKKFSDRHIVFIADRRILRKPSRKSRVKQARPRSRTLTSV 180
Query: 123 HEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGKDV 182
HE LED+V P EIVGKR +VDGSKI+KVFLD K+ + EYKL+TF +VY+ L+ KDV
Sbjct: 181 HEKYLEDLVYPTEIVGKRTTVKVDGSKIIKVFLDSKDATSLEYKLDTFKSVYKNLTSKDV 360
Query: 183 VFEYP 187
F++P
Sbjct: 361 EFKFP 375
>TC76421 homologue to PIR|T02995|T02995 unspecific monooxygenase (EC
1.14.14.1) - common tobacco, partial (44%)
Length = 5080
Score = 50.1 bits (118), Expect(2) = 5e-13
Identities = 23/24 (95%), Positives = 24/24 (99%)
Frame = +1
Query: 104 PPKKGSAAQRPRTRTLTAVHEAML 127
PPKKGSAAQRPR+RTLTAVHEAML
Sbjct: 4942 PPKKGSAAQRPRSRTLTAVHEAML 5013
Score = 40.0 bits (92), Expect(2) = 5e-13
Identities = 18/20 (90%), Positives = 19/20 (95%)
Frame = +3
Query: 129 DVVLPAEIVGKRVRYRVDGS 148
DVVLPAEIVGKR +YRVDGS
Sbjct: 5019 DVVLPAEIVGKRTKYRVDGS 5078
>BG645364
Length = 797
Score = 32.0 bits (71), Expect = 0.15
Identities = 19/36 (52%), Positives = 20/36 (54%)
Frame = -3
Query: 129 DVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTE 164
DV PA VGK RYR +GS KERNNTE
Sbjct: 102 DVTFPAVTVGKCNRYRFNGS---------KERNNTE 22
>TC88627 apyrase-like protein [Medicago truncatula]
Length = 1614
Score = 31.6 bits (70), Expect = 0.20
Identities = 12/48 (25%), Positives = 29/48 (60%)
Frame = -1
Query: 32 NTHQELKSDLKDLYINSAVQVDVSGNRKAVVIHVPYRLRKAFRKVHVK 79
N H E+++ L + + +++++ G ++ H+PYRL+ FR + ++
Sbjct: 603 NCHPEIRTFLGSINNRNCIRLNIEGT--SIAEHIPYRLQYLFRSIPIQ 466
>TC88963 similar to GP|21554778|gb|AAM63688.1 unknown {Arabidopsis
thaliana}, partial (46%)
Length = 641
Score = 30.4 bits (67), Expect = 0.44
Identities = 24/74 (32%), Positives = 34/74 (45%), Gaps = 6/74 (8%)
Frame = +1
Query: 56 GNRKAVVIH-----VPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKK-GS 109
GN +++H + Y R RK KLV E + D+ LI R + PP+K G
Sbjct: 151 GNGM*MILHQLKDSLLYSTRPEMRK---KLVEEDRAELIHSDLDLIRARMTINPPRKSGF 321
Query: 110 AAQRPRTRTLTAVH 123
A R +TR +H
Sbjct: 322 ALNRRQTRMCAMLH 363
>TC78024 similar to GP|20804963|dbj|BAB92640. hypothetical protein~similar
to Arabidopsis thaliana chromosome 3 At3g51270, partial
(43%)
Length = 1107
Score = 30.0 bits (66), Expect = 0.57
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Frame = +2
Query: 117 RTLTAVHEAMLEDVVLPAEIVGKRVRYRVDGS-KIMKVFLDPKERNNTEYKLETFAAVY 174
R LTAV M ++PAE++ + R + G+ K++K L K ++ K + F Y
Sbjct: 182 RVLTAVELGMRNHEIVPAELIDRIARLKHGGTYKVLKNLLKHKLLHHDSTKYDGFRLTY 358
>AL388681 similar to PIR|S12789|S127 GTP-binding protein ryh1 - fission
yeast (Schizosaccharomyces pombe), partial (8%)
Length = 277
Score = 29.3 bits (64), Expect = 0.98
Identities = 15/35 (42%), Positives = 21/35 (59%)
Frame = +1
Query: 23 VGQALFDLENTHQELKSDLKDLYINSAVQVDVSGN 57
+ QAL +ENT E ++ L D+ INS Q + GN
Sbjct: 58 IAQALPGMENTMNEPQNQLIDVNINSGTQNNSEGN 162
>TC90198 similar to GP|4468835|emb|CAB38221.1 hairless {Drosophila hydei},
partial (1%)
Length = 895
Score = 27.3 bits (59), Expect = 3.7
Identities = 23/80 (28%), Positives = 38/80 (46%), Gaps = 11/80 (13%)
Frame = +1
Query: 8 IHKDK---DAEPTEFEETVGQALFDLENTHQE------LKSDLKDLYINSAVQVDVSGNR 58
+H+ K DAE TE E+T FD TH+E L+ NS+ + + S N
Sbjct: 328 LHEKKQRFDAETTEVEDTFLSKGFDNAETHREDAKLDDLRKSTTSSVSNSSTEKNNSANE 507
Query: 59 K--AVVIHVPYRLRKAFRKV 76
+ ++++H P R+ + V
Sbjct: 508 ESSSLIMH-PLRVEHGTKSV 564
>TC79257 weakly similar to GP|18086351|gb|AAL57638.1 AT5g12890/T24H18_60
{Arabidopsis thaliana}, partial (27%)
Length = 1211
Score = 26.6 bits (57), Expect = 6.3
Identities = 9/14 (64%), Positives = 13/14 (92%)
Frame = -1
Query: 153 VFLDPKERNNTEYK 166
VF+DPK+ NNTE++
Sbjct: 908 VFIDPKQTNNTEFE 867
>TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog chloroplast
precursor (EC 3.4.24.-). [Alfalfa] {Medicago sativa},
complete
Length = 2473
Score = 26.6 bits (57), Expect = 6.3
Identities = 16/46 (34%), Positives = 21/46 (44%)
Frame = +3
Query: 80 LVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLTAVHEA 125
L + K KD I A RI+ P+K +A + L A HEA
Sbjct: 1665 LAARRDLKEISKDEIADALERIIAGPEKKNAVVSEEKKKLVAYHEA 1802
>TC77287 SP|Q02735|CAPP_MEDSA Phosphoenolpyruvate carboxylase (EC 4.1.1.31)
(PEPCASE). [Alfalfa] {Medicago sativa}, partial (33%)
Length = 1124
Score = 26.2 bits (56), Expect = 8.3
Identities = 18/94 (19%), Positives = 45/94 (47%), Gaps = 5/94 (5%)
Frame = +1
Query: 17 TEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKAVVIHVPYRLRKAFRKV 76
++ EET+ + +FD++ + QE+ LK+ ++ + + + + ++ R+R ++
Sbjct: 523 SDIEETLKKLVFDMKKSPQEVFDALKNQTVDLVLTAHPTQSVRRSLLQKHGRVRNCLSQL 702
Query: 77 HVKLV-----RELEKKFSGKDVILIATRRILRPP 105
+ K + +EL++ + T I R P
Sbjct: 703 YAKDITPDDKQELDEALQREIQAAFRTDEIKRTP 804
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,029,418
Number of Sequences: 36976
Number of extensions: 40458
Number of successful extensions: 214
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 212
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 213
length of query: 191
length of database: 9,014,727
effective HSP length: 91
effective length of query: 100
effective length of database: 5,649,911
effective search space: 564991100
effective search space used: 564991100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0073.3


