

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
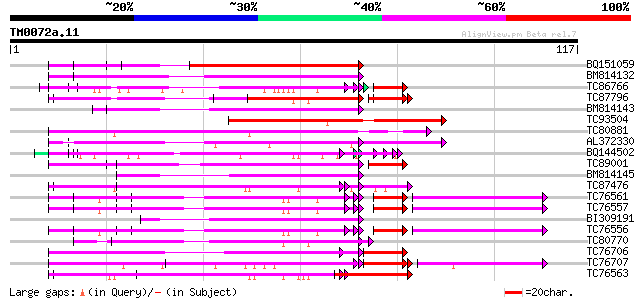
Query= TM0072a.11
(117 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 66 2e-12
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 64 8e-12
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 54 2e-10
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 58 5e-10
BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f... 57 1e-09
TC93504 weakly similar to PIR|S46286|S46286 RNA-binding protein ... 57 1e-09
TC80881 similar to PIR|T11622|T11622 extensin class 1 precursor ... 56 2e-09
AL372330 similar to GP|21322752|dbj cold shock protein-1 {Tritic... 55 3e-09
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 54 9e-09
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 54 9e-09
BM814145 homologue to GP|21322711|em pherophorin-dz1 protein {Vo... 54 1e-08
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 52 3e-08
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 50 3e-08
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 50 3e-08
BI309191 52 3e-08
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 50 3e-08
TC80770 similar to GP|4680336|gb|AAD27627.1| hypothetical protei... 50 1e-07
TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 49 3e-07
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 45 3e-07
TC76563 homologue to PIR|C29356|C29356 hydroxyproline-rich glyco... 49 4e-07
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 127 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 276
Query: 69 GGCGG 73
GG GG
Sbjct: 277 GGGGG 291
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 118 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 267
Query: 69 GGCGG 73
GG GG
Sbjct: 268 GGGGG 282
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 41 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 190
Query: 69 GGCGG 73
GG GG
Sbjct: 191GGGGG 205
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 114 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 263
Query: 69 GGCGG 73
GG GG
Sbjct: 264 GGGGG 278
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 116 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 265
Query: 69 GGCGG 73
GG GG
Sbjct: 266 GGGGG 280
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 63 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 212
Query: 69 GGCGG 73
GG GG
Sbjct: 213 GGGGG 227
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 43 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 192
Query: 69 GGCGG 73
GG GG
Sbjct: 193GGGGG 207
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 131 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 280
Query: 69 GGCGG 73
GG GG
Sbjct: 281 GGGGG 295
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 135 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 284
Query: 69 GGCGG 73
GG GG
Sbjct: 285 GGGGG 299
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 138 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 287
Query: 69 GGCGG 73
GG GG
Sbjct: 288 GGGGG 302
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 140 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 289
Query: 69 GGCGG 73
GG GG
Sbjct: 290 GGGGG 304
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 142 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 291
Query: 69 GGCGG 73
GG GG
Sbjct: 292 GGGGG 306
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 46 GGGGGGGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGG 195
Query: 69 GGCGG 73
GG GG
Sbjct: 196 GGGGG 210
Score = 63.2 bits (152), Expect = 1e-11
Identities = 32/60 (53%), Positives = 32/60 (53%)
Frame = +1
Query: 14 GGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
GG G G GG GGGG G GG GGGGGGG GGG G G GGGGG GG
Sbjct: 37 GGGGGGGGGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 186
Score = 55.1 bits (131), Expect = 4e-09
Identities = 28/53 (52%), Positives = 28/53 (52%)
Frame = +3
Query: 21 GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G GG GGGG GG GGGGGGG GGG G G GGGGG GG
Sbjct: 36 GGGGGGGGGGG-------------GGGGGGGGGGGGGGGGGGGGGGGGGGGGG 155
Score = 52.8 bits (125), Expect = 2e-08
Identities = 26/50 (52%), Positives = 28/50 (56%)
Frame = +2
Query: 24 GDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G++ GGGG GG GGGGGGG GGG G G GGGGG GG
Sbjct: 23 GENPGGGG--------------GGGGGGGGGGGGGGGGGGGGGGGGGGGG 130
Score = 52.4 bits (124), Expect = 3e-08
Identities = 22/36 (61%), Positives = 25/36 (69%)
Frame = +2
Query: 38 VMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
+ + G+ GG GGGGGGG GGG G G GGGGG GG
Sbjct: 11 IPLSGENPGGGGGGGGGGGGGGGGGGGGGGGGGGGG 118
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 63.9 bits (154), Expect = 8e-12
Identities = 32/60 (53%), Positives = 32/60 (53%)
Frame = +3
Query: 14 GGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
GG G G GG GGGG G GG GGGGGGG GGG G G GGGGG GG
Sbjct: 3 GGGRGGGGGGGGGGGGGG----------GGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 152
Score = 62.0 bits (149), Expect = 3e-11
Identities = 32/60 (53%), Positives = 32/60 (53%)
Frame = +3
Query: 14 GGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
GG G G GG GGGG G GG GGGGGGG GGG G G GGGGG GG
Sbjct: 6 GGRGGGGGGGGGGGGGGGGA-------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 164
Score = 62.0 bits (149), Expect = 3e-11
Identities = 33/65 (50%), Positives = 33/65 (50%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG A GG G G GG GGGG GG GG GGGG GGG G G GGG
Sbjct: 2 GGGAGGGGGGGGGGGGGGGGGGG-----------GGGGGGGGGRGGGGGGGGGGGGGGGG 148
Query: 69 GGCGG 73
GG GG
Sbjct: 149GGGGG 163
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 54.3 bits (129), Expect(2) = 2e-10
Identities = 32/73 (43%), Positives = 35/73 (47%), Gaps = 9/73 (12%)
Frame = -1
Query: 7 DDGGFADGGS-------SGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD-- 57
D GG DGG +GDDG GG + GGGG G GG GGGG+
Sbjct: 405 DGGGDGDGGG**GV*CGAGDDGGGGSTQGGGG----------GGGGSTQGGGDGGGGE*T 256
Query: 58 GGGNGCGSGGGGG 70
GGG C GGGGG
Sbjct: 255 GGGGECTGGGGGG 217
Score = 52.4 bits (124), Expect = 3e-08
Identities = 33/70 (47%), Positives = 34/70 (48%), Gaps = 9/70 (12%)
Frame = -1
Query: 13 DGGSSGDDG------CGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGG---GGGDGGGNGC 63
DGG GD G CG DGGGG GG GGGG GGGDGGG G
Sbjct: 405 DGGGDGDGGG**GV*CGAGDDGGGG-----------STQGGGGGGGGSTQGGGDGGG-GE 262
Query: 64 GSGGGGGCGG 73
+GGGG C G
Sbjct: 261 *TGGGGECTG 232
Score = 50.4 bits (119), Expect = 1e-07
Identities = 33/69 (47%), Positives = 35/69 (49%), Gaps = 4/69 (5%)
Frame = -1
Query: 9 GGFADGGSSGDDGCGGDSDGGG--GVVIEKVVMVEGDCSGGDSGGGGG--GGDGGGNGCG 64
GG +G + DG GGD DGGG GV C GD GGGG GG GGG G
Sbjct: 438 GGGGEG*AYTGDG-GGDGDGGG**GV*----------CGAGDDGGGGSTQGGGGGGGGST 292
Query: 65 SGGGGGCGG 73
GGG G GG
Sbjct: 291 QGGGDGGGG 265
Score = 42.0 bits (97), Expect = 3e-05
Identities = 32/85 (37%), Positives = 34/85 (39%), Gaps = 19/85 (22%)
Frame = -1
Query: 9 GGFADGGSSGDDGCG-----GDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGG-------- 55
GG GG G D G G+ GGGG E GG+ GGGGG
Sbjct: 249 GGECTGGGGGGDKYGYTGGVGE*GGGGG---------EXGGGGGE*AGGGGGEL***GTG 97
Query: 56 ------GDGGGNGCGSGGGGGCGGL 74
G GGG G G GGGG G L
Sbjct: 96 GGGENTGAGGGGGGGEFGGGGGGDL 22
Score = 40.8 bits (94), Expect = 8e-05
Identities = 32/89 (35%), Positives = 36/89 (39%), Gaps = 25/89 (28%)
Frame = -1
Query: 9 GGFADGGSS--GDDGCGGDSDGGGGVVI------EKVVMVEGDCSGGDSGGGGGGG---- 56
GG GGS+ G DG GG+ GGGG +K G G GG GGG
Sbjct: 318 GGGGGGGSTQGGGDGGGGE*TGGGGECTGGGGGGDKYGYTGGVGE*GGGGGEXGGGGGE* 139
Query: 57 -------------DGGGNGCGSGGGGGCG 72
GGG G+GGGGG G
Sbjct: 138 AGGGGGEL***GTGGGGENTGAGGGGGGG 52
Score = 38.5 bits (88), Expect(2) = 1e-05
Identities = 31/77 (40%), Positives = 33/77 (42%), Gaps = 19/77 (24%)
Frame = -1
Query: 15 GSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGG----GGGG------GDGGGNG-- 62
G S G GGD GGG EG GD GG GGGG GDGGG+G
Sbjct: 537 GPSYTGGGGGDEYTGGGG--------EGYAYTGDGGGEVYTGGGGEG*AYTGDGGGDGDG 382
Query: 63 -------CGSGGGGGCG 72
CG+G GG G
Sbjct: 381 GG**GV*CGAGDDGGGG 331
Score = 36.6 bits (83), Expect = 0.001
Identities = 26/66 (39%), Positives = 31/66 (46%), Gaps = 2/66 (3%)
Frame = -1
Query: 9 GGFADGGSSGDD--GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSG 66
GG + G G + G GG+ GGGG + G + G GGGGGG GGG G
Sbjct: 198 GGVGE*GGGGGEXGGGGGE*AGGGGGEL***GTGGGGENTGAGGGGGGGEFGGGGGGDLT 19
Query: 67 GGGGCG 72
GG G
Sbjct: 18 QYGGVG 1
Score = 28.5 bits (62), Expect(2) = 0.042
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 3/46 (6%)
Frame = -1
Query: 28 GGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCG---SGGGGG 70
GGGG E + G +GGGGG GG G G +G GGG
Sbjct: 582 GGGGDAYE--IPKTGGIGPSYTGGGGGDEYTGGGGEGYAYTGDGGG 451
Score = 25.0 bits (53), Expect = 4.4
Identities = 20/55 (36%), Positives = 22/55 (39%), Gaps = 10/55 (18%)
Frame = -1
Query: 28 GGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGG------NGCGSG----GGGGCG 72
GGGG ++ G GGGG GGG G G G GGGG G
Sbjct: 585 GGGGGDAYEIPKTGGIGPSYTGGGGGDEYTGGGGEGYAYTGDGGGEVYTGGGGEG 421
Score = 25.0 bits (53), Expect(2) = 2e-10
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = -3
Query: 76 WWRRRWW 82
WWR RWW
Sbjct: 172 WWRXRWW 152
Score = 24.3 bits (51), Expect(2) = 1e-05
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = -3
Query: 76 WWRRRWW 82
WWR RWW
Sbjct: 286 WWRWRWW 266
Score = 21.9 bits (45), Expect(2) = 0.042
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = -3
Query: 77 WRRRWWR 83
WR RWWR
Sbjct: 319 WRWRWWR 299
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 58.2 bits (139), Expect = 5e-10
Identities = 35/67 (52%), Positives = 38/67 (56%), Gaps = 2/67 (2%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD--GGGNGCGSG 66
GG+ GG SG G GG + G GV G SGG +GGG GGG GGG G GSG
Sbjct: 16 GGYGGGGGSGGGG-GGGAGGAHGV---------GYGSGGGTGGGYGGGSPGGGGGGGGSG 165
Query: 67 GGGGCGG 73
GGGG GG
Sbjct: 166GGGGGGG 186
Score = 52.8 bits (125), Expect = 2e-08
Identities = 30/64 (46%), Positives = 32/64 (49%)
Frame = +1
Query: 10 GFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGG 69
G+ GG +G G GG S GGGG GG SGGGGGGG G G G GG
Sbjct: 85 GYGSGGGTGG-GYGGGSPGGGG-------------GGGGSGGGGGGGGAHGGAYGGGIGG 222
Query: 70 GCGG 73
G GG
Sbjct: 223 GEGG 234
Score = 40.8 bits (94), Expect(2) = 6e-08
Identities = 20/36 (55%), Positives = 21/36 (57%), Gaps = 5/36 (13%)
Frame = +1
Query: 43 DCSGGDSGGGGGGGDGGG-----NGCGSGGGGGCGG 73
D GG GGGG GG GGG +G G G GGG GG
Sbjct: 7 DHGGGYGGGGGSGGGGGGGAGGAHGVGYGSGGGTGG 114
Score = 38.1 bits (87), Expect(2) = 8e-06
Identities = 16/24 (66%), Positives = 16/24 (66%)
Frame = +1
Query: 50 GGGGGGGDGGGNGCGSGGGGGCGG 73
G GGG GGG G G GGGGG GG
Sbjct: 1 GRDHGGGYGGGGGSGGGGGGGAGG 72
Score = 36.6 bits (83), Expect = 0.001
Identities = 16/27 (59%), Positives = 16/27 (59%)
Frame = +1
Query: 46 GGDSGGGGGGGDGGGNGCGSGGGGGCG 72
G D GGG GGG G G G G G GG G
Sbjct: 1 GRDHGGGYGGGGGSGGGGGGGAGGAHG 81
Score = 30.0 bits (66), Expect(2) = 6e-08
Identities = 8/9 (88%), Positives = 9/9 (99%)
Frame = +3
Query: 75 MWWRRRWWR 83
+WWRRRWWR
Sbjct: 159 LWWRRRWWR 185
Score = 25.4 bits (54), Expect(2) = 8e-06
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = +3
Query: 76 WWRRRWW 82
WWR RWW
Sbjct: 96 WWRNRWW 116
>BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (4%)
Length = 132
Score = 57.0 bits (136), Expect = 1e-09
Identities = 29/53 (54%), Positives = 29/53 (54%)
Frame = +3
Query: 21 GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G GG GGGG G GG GGGGGGG GGG G G GGGGG GG
Sbjct: 3 GGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 131
Score = 57.0 bits (136), Expect = 1e-09
Identities = 29/53 (54%), Positives = 29/53 (54%)
Frame = +1
Query: 21 GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G GG GGGG G GG GGGGGGG GGG G G GGGGG GG
Sbjct: 4 GGGGGGGGGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 132
Score = 56.2 bits (134), Expect = 2e-09
Identities = 29/56 (51%), Positives = 29/56 (51%)
Frame = +2
Query: 18 GDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G G GG GGGG GG GGGGGGG GGG G G GGGGG GG
Sbjct: 2 GGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGGGGGGGGGG 127
>TC93504 weakly similar to PIR|S46286|S46286 RNA-binding protein - wood
tobacco, partial (55%)
Length = 576
Score = 57.0 bits (136), Expect = 1e-09
Identities = 28/48 (58%), Positives = 30/48 (62%), Gaps = 3/48 (6%)
Frame = +3
Query: 46 GGDSGGGGGGGDGGGNGCG---SGGGGGCGGLMWWRRRWWRVVVVTST 90
GG GGGGG G GGG G G SGGGG GG WWR+ WWR + V T
Sbjct: 399 GGGYGGGGGYGGGGGYGGGGGYSGGGGNYGG--WWRKLWWRKLWVAET 536
Score = 25.4 bits (54), Expect = 3.3
Identities = 16/34 (47%), Positives = 17/34 (49%)
Frame = +1
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVVEGDCGDG 117
V VV VA A +VVVV VVVE G G
Sbjct: 397 VAVVMVAAVAMAAAVAMVVVVAIVVVVEIMVGGG 498
Score = 23.9 bits (50), Expect = 9.7
Identities = 6/9 (66%), Positives = 7/9 (77%)
Frame = +2
Query: 75 MWWRRRWWR 83
+WWR WWR
Sbjct: 392 VWWRWLWWR 418
>TC80881 similar to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (34%)
Length = 597
Score = 56.2 bits (134), Expect = 2e-09
Identities = 39/101 (38%), Positives = 46/101 (44%), Gaps = 22/101 (21%)
Frame = -3
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGD---------------- 48
GG +G G D G GG+ +GGGG + EGD GGD
Sbjct: 583 GGGGEGDGGGGDL*TYGGGGEGEGGGGDLYT*FGGSEGDGGGGDL*TYGGGGDGEGGGGL 404
Query: 49 --SGGGGGGGDGGGNGCGSGGGGGCGGLMWWRRRWWRVVVV 87
+ GGGG GDGGG + GGGG G WWR WR VV+
Sbjct: 403 L*TYGGGGEGDGGGGDL*TYGGGGEG--RWWR---WRFVVI 296
Score = 30.4 bits (67), Expect = 0.10
Identities = 20/47 (42%), Positives = 22/47 (46%), Gaps = 12/47 (25%)
Frame = -3
Query: 40 VEGDCSGGDSGGG-----GGGGDGGGNG-------CGSGGGGGCGGL 74
+ G GD GGG GGGG+G G G GS G GG G L
Sbjct: 589 IYGGGGEGDGGGGDL*TYGGGGEGEGGGGDLYT*FGGSEGDGGGGDL 449
Score = 30.0 bits (66), Expect = 0.14
Identities = 21/61 (34%), Positives = 27/61 (43%), Gaps = 23/61 (37%)
Frame = -1
Query: 39 MVEGDCSGGD---------SGGGGGGGDGGGNG--------------CGSGGGGGCGGLM 75
+V+GD GGD + GGGG G+GGG C GG GCG L+
Sbjct: 336 VVKGDGGGGDL***GGYLWTYGGGGDGEGGGGDL**YGGDLK*YGGCCP*LGGVGCG*LL 157
Query: 76 W 76
+
Sbjct: 156 F 154
Score = 28.5 bits (62), Expect = 0.39
Identities = 17/35 (48%), Positives = 18/35 (50%), Gaps = 4/35 (11%)
Frame = -1
Query: 1 MVAVVVDDGGFAD----GGSSGDDGCGGDSDGGGG 31
M VV DGG D GG G GGD +GGGG
Sbjct: 345 MEEVVKGDGGGGDL***GGYLWTYGGGGDGEGGGG 241
>AL372330 similar to GP|21322752|dbj cold shock protein-1 {Triticum
aestivum}, partial (33%)
Length = 435
Score = 55.5 bits (132), Expect = 3e-09
Identities = 33/65 (50%), Positives = 34/65 (51%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG+ GGSSG G GGGG G SGG GGG GGG GGG G G GG
Sbjct: 122 GGY--GGSSGGYGGSSGGYGGGGY--------GGGASGGYGGGGYGGGSGGGYGGGGYGG 271
Query: 69 GGCGG 73
GG GG
Sbjct: 272 GGYGG 286
Score = 45.1 bits (105), Expect = 4e-06
Identities = 32/84 (38%), Positives = 38/84 (45%), Gaps = 2/84 (2%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGG-GGGGDGGGNGCGSGG 67
GG+ GG G G G GGG G GG GGG GGGG GGG G G GG
Sbjct: 164 GGYGGGGYGG--GASGGYGGGG---------YGGGSGGGYGGGGYGGGGYGGGGGGGYGG 310
Query: 68 GG-GCGGLMWWRRRWWRVVVVTST 90
G G ++ W R+ ++ T
Sbjct: 311 DKMGALGANLQKQDWGRIELIPFT 382
Score = 43.5 bits (101), Expect = 1e-05
Identities = 27/64 (42%), Positives = 27/64 (42%), Gaps = 3/64 (4%)
Frame = +2
Query: 13 DGGSSGDDGCGGDSDGGGGVVIEKVVMVE---GDCSGGDSGGGGGGGDGGGNGCGSGGGG 69
D SS G GG S GG G G GG SGG GG G G G GGG
Sbjct: 2 DYRSSSSGGYGGSSSGGYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGGYGGSSGGYGGG 181
Query: 70 GCGG 73
G GG
Sbjct: 182GYGG 193
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 53.9 bits (128), Expect = 9e-09
Identities = 33/72 (45%), Positives = 34/72 (46%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG+ GG G G GGGG G C GG GGGGGG GG G GGG
Sbjct: 57 GGWRRGGEVGGSGWCVWGGGGGG----------GRCGGG---GGGGGGRGGAEAAGPGGG 197
Query: 69 GGCGGLMWWRRR 80
GG GG RRR
Sbjct: 198 GGGGGDALGRRR 233
Score = 47.4 bits (111), Expect = 8e-07
Identities = 33/78 (42%), Positives = 34/78 (43%), Gaps = 13/78 (16%)
Frame = +3
Query: 14 GGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD---------GGGNGCG 64
GG G CGG GGGG G + G GGGGGGGD G G G G
Sbjct: 108 GGGGGGGRCGGGGGGGGGR--------GGAEAAGPGGGGGGGGDALGRRRLAKGSGAGGG 263
Query: 65 SG----GGGGCGGLMWWR 78
G GGGG GG W R
Sbjct: 264 GGARSRGGGGPGG--WGR 311
Score = 43.5 bits (101), Expect = 1e-05
Identities = 31/66 (46%), Positives = 35/66 (52%), Gaps = 9/66 (13%)
Frame = +2
Query: 13 DGGS-------SGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDG--GGNGC 63
DGGS G G GG + GGGG + ++V V G GG GG GGG G GG G
Sbjct: 5 DGGSY*LYTRRGGGAGGGGLAAGGGGGGL-RLVRVGGWGRGGPLRGGWGGGRGPRGGGGG 181
Query: 64 GSGGGG 69
G GGGG
Sbjct: 182GPGGGG 199
Score = 42.7 bits (99), Expect = 2e-05
Identities = 25/69 (36%), Positives = 32/69 (46%), Gaps = 1/69 (1%)
Frame = +3
Query: 9 GGFADGGSSGDDGCG-GDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGG 67
GG GG G + G G GGGG + + + + G +GGGGG GG G G G
Sbjct: 141 GGGGGGGRGGAEAAGPGGGGGGGGDALGRRRLAK----GSGAGGGGGARSRGGGGPGGWG 308
Query: 68 GGGCGGLMW 76
G G +W
Sbjct: 309 RAGQGTGLW 335
Score = 39.3 bits (90), Expect = 2e-04
Identities = 30/87 (34%), Positives = 31/87 (35%), Gaps = 22/87 (25%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDG---------- 58
G A GG G G GGGG G GG +GGGG GG G
Sbjct: 424 GSGALGGRRGARGRAPRGGGGGGGAASDCGGALGGDGGGGTGGGGRGGRGYVATRTRGTS 603
Query: 59 GGNGC------------GSGGGGGCGG 73
G GC GGGGG GG
Sbjct: 604 AGRGCPPIFLARGTRSHSGGGGGGRGG 684
Score = 38.5 bits (88), Expect = 4e-04
Identities = 29/69 (42%), Positives = 31/69 (44%), Gaps = 2/69 (2%)
Frame = +3
Query: 15 GSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGGL 74
G GD GG GG + EG G +GGG GGG G G GSGGG G G
Sbjct: 378 GHHGDALTGGARVAGG---VGGAGGPEGGARAGAAGGGRGGGRGLGLWRGSGGGRGRGD- 545
Query: 75 MWWR--RRW 81
WR RW
Sbjct: 546 --WRGGARW 566
Score = 37.7 bits (86), Expect = 6e-04
Identities = 27/71 (38%), Positives = 28/71 (39%), Gaps = 4/71 (5%)
Frame = +1
Query: 6 VDDGGFADGGSSGDDGCGGD----SDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGN 61
V GG A GG G G G + GGG V G G G GGG G G
Sbjct: 112 VGAGGAAAGGVGGGAGAEGGRRRRARGGGAVAGGMPSAAGGLLRGVGPGAGGGPGPGAAG 291
Query: 62 GCGSGGGGGCG 72
G GGG G G
Sbjct: 292 GRVGGGGRGRG 324
Score = 37.4 bits (85), Expect = 8e-04
Identities = 29/81 (35%), Positives = 32/81 (38%), Gaps = 17/81 (20%)
Frame = +1
Query: 9 GGFAD--GGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSG-------------GDSGGGG 53
GG A GG+ G DG GG GG G G +G SGGGG
Sbjct: 490 GGAASDCGGALGGDGGGGTGGGGRGGRGYVATRTRGTSAGRGCPPIFLARGTRSHSGGGG 669
Query: 54 GGGDGGGNGCGSGGGGG--CG 72
GG GG G +G G CG
Sbjct: 670 GGRGGGRRGVAAGCARGMLCG 732
Score = 36.6 bits (83), Expect = 0.001
Identities = 26/71 (36%), Positives = 29/71 (40%), Gaps = 6/71 (8%)
Frame = +1
Query: 11 FADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSG---- 66
+ GG G G GG GGG + G C G +GG GG GGG G G
Sbjct: 28 YQKGGGRGGRGAGG----GGG----RWGAQAGACGGVGAGGAAAGGVGGGAGAEGGRRRR 183
Query: 67 --GGGGCGGLM 75
GGG G M
Sbjct: 184ARGGGAVAGGM 216
Score = 33.1 bits (74), Expect = 0.016
Identities = 33/107 (30%), Positives = 38/107 (34%), Gaps = 35/107 (32%)
Frame = +2
Query: 8 DGGFADG-------GSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGG 60
DGG A G G+ G G G GG ++V EG G +GG GGG+ GG
Sbjct: 398 DGGRAGGRRGRGRWGAGGGRAGGRRGGGAGGGARPRIV--EGLWGGTGAGGLAGGGEVGG 571
Query: 61 N----------------------------GCGSGGGGGCGGLMWWRR 79
G GGGGG GG WRR
Sbjct: 572 GM*RLGREGRAPGGAAHQFSWQEGRVPIPGGEEGGGGGGGG--GWRR 706
Score = 31.6 bits (70), Expect = 0.047
Identities = 19/54 (35%), Positives = 22/54 (40%), Gaps = 10/54 (18%)
Frame = +1
Query: 37 VVMVEGDCSGGDSGGGGGG----------GDGGGNGCGSGGGGGCGGLMWWRRR 80
++ +G GG GGGGG G G G G GGG G RRR
Sbjct: 22 IIYQKGGGRGGRGAGGGGGRWGAQAGACGGVGAGGAAAGGVGGGAGAEGGRRRR 183
Score = 31.2 bits (69), Expect = 0.061
Identities = 28/87 (32%), Positives = 34/87 (38%), Gaps = 15/87 (17%)
Frame = +2
Query: 15 GSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDS-------------GGGGGGGDGGGN 61
G +G G G + GGG+ + EG GG + GG GGG GGG
Sbjct: 524 GGTGAGGLAGGGEVGGGM*R---LGREGRAPGGAAHQFSWQEGRVPIPGGEEGGGGGGGG 694
Query: 62 GCGSGGGGGCGGLMWWRRR--WWRVVV 86
G G GC + R R WR V
Sbjct: 695 GWRRGAREGCCAALARRVRLGGWRWAV 775
Score = 28.9 bits (63), Expect = 0.30
Identities = 26/85 (30%), Positives = 28/85 (32%), Gaps = 21/85 (24%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGG------------GGGG 56
GG GG G G + GG G G + G GGG GGG
Sbjct: 49 GGRGAGGGGGRWGAQAGACGGVGA---------GGAAAGGVGGGAGAEGGRRRRARGGGA 201
Query: 57 DGGG---------NGCGSGGGGGCG 72
GG G G G GGG G
Sbjct: 202 VAGGMPSAAGGLLRGVGPGAGGGPG 276
Score = 28.5 bits (62), Expect = 0.39
Identities = 17/40 (42%), Positives = 17/40 (42%), Gaps = 13/40 (32%)
Frame = +3
Query: 46 GGDSGGGGGGGDGGGN------------GCGSGGGGG-CG 72
GG G GGG GGG GC GGG G CG
Sbjct: 660 GGRRGAGGGEEGGGGGVRARDAVRRSRAGCAWGGGAGRCG 779
Score = 28.5 bits (62), Expect = 0.39
Identities = 18/51 (35%), Positives = 19/51 (36%), Gaps = 19/51 (37%)
Frame = +3
Query: 41 EGDCSGGDSGGGG-------------------GGGDGGGNGCGSGGGGGCG 72
EG + GGG GG GGG CG GGGGG G
Sbjct: 9 EGAINYIPEGGGARGAGGWRRGGEVGGSGWCVWGGGGGGGRCGGGGGGGGG 161
Score = 26.9 bits (58), Expect = 1.1
Identities = 15/29 (51%), Positives = 15/29 (51%)
Frame = +2
Query: 54 GGGDGGGNGCGSGGGGGCGGLMWWRRRWW 82
GGG GGG G GGG GGL R W
Sbjct: 38 GGGAGGG---GLAAGGGGGGLRLVRVGGW 115
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 53.9 bits (128), Expect = 9e-09
Identities = 31/65 (47%), Positives = 31/65 (47%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG D SGGGGG G GG G G GGG
Sbjct: 318 GGGGYGGGGGYGGGGGGYGGGGG---------RRDGGYSRSGGGGGYGGGGDRGYGGGGG 470
Query: 69 GGCGG 73
GG GG
Sbjct: 471 GGYGG 485
Score = 45.4 bits (106), Expect(2) = 8e-07
Identities = 26/51 (50%), Positives = 27/51 (51%)
Frame = +3
Query: 23 GGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G D DG V E + SGG GGGGGG GGG G G GGGG GG
Sbjct: 240 GQDIDGRNITVNE----AQSRGSGGGGRGGGGGGYGGGGGYGGGGGGYGGG 380
Score = 38.1 bits (87), Expect = 5e-04
Identities = 24/53 (45%), Positives = 24/53 (45%)
Frame = +3
Query: 21 GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G GG GGGG G GGGGG G GGG G G GGG GG
Sbjct: 291 GSGGGGRGGGG---------------GGYGGGGGYG-GGGGGYGGGGGRRDGG 401
Score = 31.6 bits (70), Expect = 0.047
Identities = 19/53 (35%), Positives = 21/53 (38%)
Frame = +3
Query: 8 DGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGG 60
DGG++ G G G GGD GGGGGGG GGG
Sbjct: 393 DGGYSRSGGGGGYGGGGDR---------------------GYGGGGGGGYGGG 488
Score = 26.2 bits (56), Expect = 2.0
Identities = 15/26 (57%), Positives = 16/26 (60%)
Frame = +1
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVV 109
VVVV V AVV V VV+ EAVV
Sbjct: 289 VVVVEEVEVVAAVVMVAAVVMEEAVV 366
Score = 21.6 bits (44), Expect(2) = 8e-07
Identities = 5/8 (62%), Positives = 6/8 (74%)
Frame = +2
Query: 75 MWWRRRWW 82
+WW R WW
Sbjct: 452 IWWWRWWW 475
>BM814145 homologue to GP|21322711|em pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (3%)
Length = 111
Score = 53.5 bits (127), Expect = 1e-08
Identities = 27/51 (52%), Positives = 27/51 (52%)
Frame = +1
Query: 23 GGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
GG GGGG GG GGGGGGG GGG G G GGGGG GG
Sbjct: 1 GGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGGGGGGGGGG 111
Score = 36.6 bits (83), Expect = 0.001
Identities = 20/50 (40%), Positives = 21/50 (42%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDG 58
GG+ GG G G GG GGGG GGGGGGG G
Sbjct: 15 GGWGGGGGGGGGGGGGGGGGGGG------------------GGGGGGGGG 110
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 52.4 bits (124), Expect = 3e-08
Identities = 42/106 (39%), Positives = 48/106 (44%), Gaps = 31/106 (29%)
Frame = -3
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGD--SGGGGGGGDG---- 58
GG +G G D G GGD +GGGG + E + GGD + GGGG GDG
Sbjct: 634 GGGGEGEGGGGDL*PYGGGGDGEGGGGDL**YGGGGEAEGGGGDL*T*GGGGEGDGGGGD 455
Query: 59 ----GGNGCGSGGGG----------GCGGL-----MW--WRRRWWR 83
GG G G GGGG G GG +W W RRWWR
Sbjct: 454 L*TYGGGGEGEGGGGDL*TYGGGGEGDGGRW*LVDIWRRW*RRWWR 317
Score = 49.7 bits (117), Expect = 2e-07
Identities = 34/75 (45%), Positives = 36/75 (47%), Gaps = 14/75 (18%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGD----SGGGGGGGDG-- 58
GG +G G D G GGD DGGGG + EGD GGD GGG G G G
Sbjct: 345 GGGGEGDGGGGDL*TYGGGGDGDGGGGDL*TYGGGGEGDGGGGDL*I*GGGGEGVGGGGD 166
Query: 59 ----GGNGCGSGGGG 69
GG G G GGGG
Sbjct: 165 L*TYGGGGDGEGGGG 121
Score = 48.5 bits (114), Expect = 4e-07
Identities = 30/67 (44%), Positives = 36/67 (52%), Gaps = 6/67 (8%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGD--SGGGGGGGDGGGNG 62
GG +G G D G GGD +GGGG++ +GD GGD + GGGG GDGGG
Sbjct: 201 GGGGEGVGGGGDL*TYGGGGDGEGGGGLL*TYGGGGDGDGGGGDLYTYGGGGEGDGGGGD 22
Query: 63 CGSGGGG 69
GGG
Sbjct: 21 LYIYGGG 1
Score = 45.4 bits (106), Expect = 3e-06
Identities = 33/75 (44%), Positives = 35/75 (46%), Gaps = 14/75 (18%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGD----SGGGGGGGDG-- 58
GG DG G D G GG+ DGGGG + EG GGD GGG G G G
Sbjct: 297 GGGGDGDGGGGDL*TYGGGGEGDGGGGDL*I*GGGGEGVGGGGDL*TYGGGGDGEGGGGL 118
Query: 59 ----GGNGCGSGGGG 69
GG G G GGGG
Sbjct: 117 L*TYGGGGDGDGGGG 73
Score = 43.5 bits (101), Expect = 1e-05
Identities = 29/68 (42%), Positives = 32/68 (46%), Gaps = 4/68 (5%)
Frame = -1
Query: 10 GFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGD----SGGGGGGGDGGGNGCGS 65
G GG G GG+ DGGGG + +GD GGD GGG G G GG
Sbjct: 378 GMVVGGDL*IYGGGGEGDGGGGDL*TYGGGGDGDGGGGDL*TYGGGGEGDGGGGDL*I*G 199
Query: 66 GGGGGCGG 73
GGG G GG
Sbjct: 198 GGGEGVGG 175
Score = 40.8 bits (94), Expect = 8e-05
Identities = 25/50 (50%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Frame = -3
Query: 23 GGDSDGGGGVVIEKVVMVEGDCSGGD--SGGGGGGGDGGGNGCGSGGGGG 70
GGD DGGGG + EG+ GGD GGGG G+GGG GGGG
Sbjct: 676 GGDGDGGGGDL*I*GGGGEGEGGGGDL*PYGGGGDGEGGGGDL**YGGGG 527
Score = 39.3 bits (90), Expect = 2e-04
Identities = 29/75 (38%), Positives = 34/75 (44%), Gaps = 14/75 (18%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGG----DSGGGGGGGDG-- 58
GG +G G D G GG+ GGGG + +G+ GG GGG G G G
Sbjct: 249 GGGGEGDGGGGDL*I*GGGGEGVGGGGDL*TYGGGGDGEGGGGLL*TYGGGGDGDGGGGD 70
Query: 59 ----GGNGCGSGGGG 69
GG G G GGGG
Sbjct: 69 LYTYGGGGEGDGGGG 25
Score = 35.8 bits (81), Expect = 0.002
Identities = 20/37 (54%), Positives = 23/37 (62%), Gaps = 2/37 (5%)
Frame = -1
Query: 38 VMVEGDCSGGDS--GGGGGGGDGGGNGCGSGGGGGCG 72
V+V+G GGD GGGG GDGGG + GGGG G
Sbjct: 390 VVVKGMVVGGDL*IYGGGGEGDGGGGDL*TYGGGGDG 280
Score = 28.1 bits (61), Expect(2) = 0.009
Identities = 14/25 (56%), Positives = 14/25 (56%), Gaps = 6/25 (24%)
Frame = -3
Query: 52 GGGGGDGGG------NGCGSGGGGG 70
GGG GDGGG G G G GGG
Sbjct: 679 GGGDGDGGGGDL*I*GGGGEGEGGG 605
Score = 27.7 bits (60), Expect = 0.67
Identities = 14/26 (53%), Positives = 14/26 (53%), Gaps = 6/26 (23%)
Frame = -3
Query: 50 GGGGGGGDG------GGNGCGSGGGG 69
GGG G G G GG G G GGGG
Sbjct: 679 GGGDGDGGGGDL*I*GGGGEGEGGGG 602
Score = 24.6 bits (52), Expect = 5.7
Identities = 14/41 (34%), Positives = 18/41 (43%), Gaps = 6/41 (14%)
Frame = -2
Query: 76 WWRRRWWRVV------VVTSTVVATAVVTVVVVVVVEAVVV 110
WW R WW VV V +V + + VV +E V V
Sbjct: 389 WW*RGWWSVVTCRYMEAVVKEMVEEEIYKRMEVVGMETVEV 267
Score = 24.6 bits (52), Expect(2) = 0.009
Identities = 10/17 (58%), Positives = 11/17 (63%), Gaps = 4/17 (23%)
Frame = -2
Query: 75 MWWRRRW----WRVVVV 87
+WWRR W WR VVV
Sbjct: 590 IWWRR*WRRWRWRFVVV 540
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 50.4 bits (119), Expect(2) = 3e-08
Identities = 30/61 (49%), Positives = 31/61 (50%), Gaps = 4/61 (6%)
Frame = +1
Query: 14 GGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGG----DGGGNGCGSGGGG 69
GG G G GG GGGG E+ G GG GGGGGGG GGG G GGGG
Sbjct: 604 GGGGGGRGGGGYGGGGGGYGGER----RGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGG 771
Query: 70 G 70
G
Sbjct: 772 G 774
Score = 43.9 bits (102), Expect = 9e-06
Identities = 29/68 (42%), Positives = 31/68 (44%), Gaps = 6/68 (8%)
Frame = +1
Query: 9 GGFADGGSS------GDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNG 62
GG+ GG G G GG GGGG E+ GG S GGGGGG GGG
Sbjct: 631 GGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGER-----RGGGGGYSRGGGGGGYGGGGY 795
Query: 63 CGSGGGGG 70
GG GG
Sbjct: 796 SRGGGDGG 819
Score = 43.5 bits (101), Expect(2) = 2e-06
Identities = 26/54 (48%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Frame = +1
Query: 23 GGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNG---CGSGGGGGCGG 73
G D DG V + G GG GGGG GG GGG G G GGGGG GG
Sbjct: 544 GQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGG 705
Score = 43.5 bits (101), Expect(2) = 3e-06
Identities = 26/53 (49%), Positives = 26/53 (49%), Gaps = 6/53 (11%)
Frame = +1
Query: 26 SDGGGGVVIEKVVMVEGDCSGGDSGGGGGG------GDGGGNGCGSGGGGGCG 72
S GGGG G GG GGGGGG G GGG G G GGGGG G
Sbjct: 598 SGGGGG----------GGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYG 726
Score = 34.3 bits (77), Expect = 0.007
Identities = 19/31 (61%), Positives = 19/31 (61%)
Frame = +1
Query: 49 SGGGGGGGDGGGNGCGSGGGGGCGGLMWWRR 79
SGGGGGGG GGG G GGG GG RR
Sbjct: 598 SGGGGGGGRGGG-----GYGGGGGGYGGERR 675
Score = 21.9 bits (45), Expect(2) = 4.9
Identities = 13/26 (50%), Positives = 15/26 (57%)
Frame = +2
Query: 86 VVTSTVVATAVVTVVVVVVVEAVVVE 111
VV +V VVTV VV E VVV+
Sbjct: 644 VVAVDMVENVVVTVEVVDTEEEVVVD 721
Score = 21.9 bits (45), Expect(2) = 2e-06
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = +2
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVVE 111
VVV V AV T VVV + VVV+
Sbjct: 710 VVVDMENAVVEAVATAEEVVVEDMVVVD 793
Score = 21.2 bits (43), Expect(2) = 3e-06
Identities = 5/7 (71%), Positives = 5/7 (71%)
Frame = +3
Query: 76 WWRRRWW 82
WW R WW
Sbjct: 765 WWWRIWW 785
Score = 21.2 bits (43), Expect(2) = 3e-08
Identities = 5/7 (71%), Positives = 5/7 (71%)
Frame = +3
Query: 76 WWRRRWW 82
WWR WW
Sbjct: 768 WWRIWWW 788
Score = 21.2 bits (43), Expect(2) = 4.9
Identities = 6/8 (75%), Positives = 6/8 (75%)
Frame = +3
Query: 76 WWRRRWWR 83
W RR WWR
Sbjct: 609 WRRRPWWR 632
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 50.4 bits (119), Expect(2) = 3e-08
Identities = 30/61 (49%), Positives = 31/61 (50%), Gaps = 4/61 (6%)
Frame = +1
Query: 14 GGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGG----DGGGNGCGSGGGG 69
GG G G GG GGGG E+ G GG GGGGGGG GGG G GGGG
Sbjct: 508 GGGGGGRGGGGYGGGGGGYGGER----RGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGG 675
Query: 70 G 70
G
Sbjct: 676 G 678
Score = 43.9 bits (102), Expect = 9e-06
Identities = 29/68 (42%), Positives = 31/68 (44%), Gaps = 6/68 (8%)
Frame = +1
Query: 9 GGFADGGSS------GDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNG 62
GG+ GG G G GG GGGG E+ GG S GGGGGG GGG
Sbjct: 535 GGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGER-----RGGGGGYSRGGGGGGYGGGGY 699
Query: 63 CGSGGGGG 70
GG GG
Sbjct: 700 SRGGGDGG 723
Score = 43.5 bits (101), Expect(2) = 2e-06
Identities = 26/54 (48%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Frame = +1
Query: 23 GGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNG---CGSGGGGGCGG 73
G D DG V + G GG GGGG GG GGG G G GGGGG GG
Sbjct: 448 GQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGG 609
Score = 43.5 bits (101), Expect(2) = 3e-06
Identities = 26/53 (49%), Positives = 26/53 (49%), Gaps = 6/53 (11%)
Frame = +1
Query: 26 SDGGGGVVIEKVVMVEGDCSGGDSGGGGGG------GDGGGNGCGSGGGGGCG 72
S GGGG G GG GGGGGG G GGG G G GGGGG G
Sbjct: 502 SGGGGG----------GGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYG 630
Score = 34.3 bits (77), Expect = 0.007
Identities = 19/31 (61%), Positives = 19/31 (61%)
Frame = +1
Query: 49 SGGGGGGGDGGGNGCGSGGGGGCGGLMWWRR 79
SGGGGGGG GGG G GGG GG RR
Sbjct: 502 SGGGGGGGRGGG-----GYGGGGGGYGGERR 579
Score = 21.9 bits (45), Expect(2) = 4.9
Identities = 13/26 (50%), Positives = 15/26 (57%)
Frame = +2
Query: 86 VVTSTVVATAVVTVVVVVVVEAVVVE 111
VV +V VVTV VV E VVV+
Sbjct: 548 VVAVDMVENVVVTVEVVDTEEEVVVD 625
Score = 21.9 bits (45), Expect(2) = 2e-06
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = +2
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVVE 111
VVV V AV T VVV + VVV+
Sbjct: 614 VVVDMENAVVEAVATAEEVVVEDMVVVD 697
Score = 21.2 bits (43), Expect(2) = 3e-06
Identities = 5/7 (71%), Positives = 5/7 (71%)
Frame = +3
Query: 76 WWRRRWW 82
WW R WW
Sbjct: 669 WWWRIWW 689
Score = 21.2 bits (43), Expect(2) = 3e-08
Identities = 5/7 (71%), Positives = 5/7 (71%)
Frame = +3
Query: 76 WWRRRWW 82
WWR WW
Sbjct: 672 WWRIWWW 692
Score = 21.2 bits (43), Expect(2) = 4.9
Identities = 6/8 (75%), Positives = 6/8 (75%)
Frame = +3
Query: 76 WWRRRWWR 83
W RR WWR
Sbjct: 513 WRRRPWWR 536
>BI309191
Length = 109
Score = 52.0 bits (123), Expect = 3e-08
Identities = 26/46 (56%), Positives = 26/46 (56%)
Frame = +2
Query: 28 GGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
GGGG G GG GGGGGGG GGG G G GGGGG GG
Sbjct: 2 GGGG----------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 109
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 50.4 bits (119), Expect(2) = 3e-08
Identities = 30/61 (49%), Positives = 31/61 (50%), Gaps = 4/61 (6%)
Frame = +2
Query: 14 GGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGG----DGGGNGCGSGGGG 69
GG G G GG GGGG E+ G GG GGGGGGG GGG G GGGG
Sbjct: 350 GGGGGGRGGGGYGGGGGGYGGER----RGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGG 517
Query: 70 G 70
G
Sbjct: 518 G 520
Score = 43.9 bits (102), Expect = 9e-06
Identities = 29/68 (42%), Positives = 31/68 (44%), Gaps = 6/68 (8%)
Frame = +2
Query: 9 GGFADGGSS------GDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNG 62
GG+ GG G G GG GGGG E+ GG S GGGGGG GGG
Sbjct: 377 GGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGER-----RGGGGGYSRGGGGGGYGGGGY 541
Query: 63 CGSGGGGG 70
GG GG
Sbjct: 542 SRGGGDGG 565
Score = 43.5 bits (101), Expect(2) = 2e-06
Identities = 26/54 (48%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Frame = +2
Query: 23 GGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNG---CGSGGGGGCGG 73
G D DG V + G GG GGGG GG GGG G G GGGGG GG
Sbjct: 290 GQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGG 451
Score = 43.5 bits (101), Expect(2) = 3e-06
Identities = 26/53 (49%), Positives = 26/53 (49%), Gaps = 6/53 (11%)
Frame = +2
Query: 26 SDGGGGVVIEKVVMVEGDCSGGDSGGGGGG------GDGGGNGCGSGGGGGCG 72
S GGGG G GG GGGGGG G GGG G G GGGGG G
Sbjct: 344 SGGGGG----------GGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYG 472
Score = 34.3 bits (77), Expect = 0.007
Identities = 19/31 (61%), Positives = 19/31 (61%)
Frame = +2
Query: 49 SGGGGGGGDGGGNGCGSGGGGGCGGLMWWRR 79
SGGGGGGG GGG G GGG GG RR
Sbjct: 344 SGGGGGGGRGGG-----GYGGGGGGYGGERR 421
Score = 21.9 bits (45), Expect(2) = 5.0
Identities = 13/26 (50%), Positives = 15/26 (57%)
Frame = +3
Query: 86 VVTSTVVATAVVTVVVVVVVEAVVVE 111
VV +V VVTV VV E VVV+
Sbjct: 390 VVAVDMVENVVVTVEVVDTEEEVVVD 467
Score = 21.9 bits (45), Expect(2) = 2e-06
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = +3
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVVE 111
VVV V AV T VVV + VVV+
Sbjct: 456 VVVDMENAVVEAVATAEEVVVEDMVVVD 539
Score = 21.2 bits (43), Expect(2) = 3e-06
Identities = 5/7 (71%), Positives = 5/7 (71%)
Frame = +1
Query: 76 WWRRRWW 82
WWR WW
Sbjct: 514 WWRIWWW 534
Score = 21.2 bits (43), Expect(2) = 3e-08
Identities = 5/7 (71%), Positives = 5/7 (71%)
Frame = +1
Query: 76 WWRRRWW 82
WW R WW
Sbjct: 511 WWWRIWW 531
Score = 21.2 bits (43), Expect(2) = 5.0
Identities = 6/8 (75%), Positives = 6/8 (75%)
Frame = +1
Query: 76 WWRRRWWR 83
W RR WWR
Sbjct: 355 WRRRPWWR 378
>TC80770 similar to GP|4680336|gb|AAD27627.1| hypothetical protein {Oryza
sativa subsp. indica} [Oryza sativa (indica
cultivar-group)], partial (7%)
Length = 507
Score = 50.4 bits (119), Expect = 1e-07
Identities = 27/64 (42%), Positives = 32/64 (49%), Gaps = 10/64 (15%)
Frame = +1
Query: 22 CGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGG----------GDGGGNGCGSGGGGGC 71
C G GG G +I+ G C GG GG GGG G G G GCG+G GGGC
Sbjct: 1 CDGGCGGGCGNIIKS-----GGCGGGCGGGCGGGCGNIVKSGGCGGGCGGGCGAGCGGGC 165
Query: 72 GGLM 75
G ++
Sbjct: 166GSMV 177
Score = 47.8 bits (112), Expect = 6e-07
Identities = 31/79 (39%), Positives = 35/79 (44%), Gaps = 19/79 (24%)
Frame = +1
Query: 14 GGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGG------------- 60
GG G GCGG GG G +++ G C GG GGG G G GGG
Sbjct: 46 GGCGG--GCGGGCGGGCGNIVKS-----GGCGGG-CGGGCGAGCGGGCGSMVGSGGCENY 201
Query: 61 ------NGCGSGGGGGCGG 73
+GCG G G GCGG
Sbjct: 202 DNMKKSSGCGEGCGEGCGG 258
Score = 28.1 bits (61), Expect = 0.51
Identities = 24/78 (30%), Positives = 28/78 (35%), Gaps = 10/78 (12%)
Frame = +1
Query: 5 VVDDGGFADG-----GSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDS-GGGGGGGDG 58
+V GG G G+ GCG GG C D+ G G+G
Sbjct: 97 IVKSGGCGGGCGGGCGAGCGGGCGSMVGSGG-------------CENYDNMKKSSGCGEG 237
Query: 59 GGNGCGSGGG----GGCG 72
G GCG GG G CG
Sbjct: 238 CGEGCGGCGGDILDGKCG 291
Score = 25.8 bits (55), Expect = 2.6
Identities = 6/8 (75%), Positives = 7/8 (87%)
Frame = +3
Query: 75 MWWRRRWW 82
+WWR RWW
Sbjct: 114 LWWRMRWW 137
>TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (94%)
Length = 1034
Score = 48.9 bits (115), Expect = 3e-07
Identities = 27/65 (41%), Positives = 33/65 (50%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG+ +GG +G GG + GGGG +GG GG GG +GGG G GG
Sbjct: 253 GGYHNGGGGYHNGGGGYNHGGGGY------------NGGGGHGGHGGYNGGGGHGGYNGG 396
Query: 69 GGCGG 73
GG GG
Sbjct: 397 GGHGG 411
Score = 38.9 bits (89), Expect = 3e-04
Identities = 24/61 (39%), Positives = 30/61 (48%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG+ +GG + G GG + GGG G G + GGG GG +GGG G GG
Sbjct: 274 GGYHNGGGGYNHGGGGYNGGGG----------HGGHGGYNGGGGHGGYNGGG---GHGGH 414
Query: 69 G 69
G
Sbjct: 415 G 417
Score = 37.0 bits (84), Expect = 0.001
Identities = 30/89 (33%), Positives = 34/89 (37%), Gaps = 23/89 (25%)
Frame = +1
Query: 8 DGGFADGGSSGDDGCGG-------------------DSDGGGGVVIEKVVMVEGDCSGGD 48
+GG GG +G G GG D+ GGG G S
Sbjct: 361 NGGGGHGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGSYNHGGSYNHGGGSYNH 540
Query: 49 SGG----GGGGGDGGGNGCGSGGGGGCGG 73
GG GGGG + GG G G GGGG GG
Sbjct: 541 GGGSYHHGGGGYNHGGGGHGGHGGGGHGG 627
Score = 33.9 bits (76), Expect(2) = 6e-05
Identities = 24/65 (36%), Positives = 27/65 (40%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG+ GG + GG +GGG GG GGGG GG G GGG
Sbjct: 187 GGYNHGGYNH----GGGYNGGGYN------------HGGGYHNGGGGYHNGGGGYNHGGG 318
Query: 69 GGCGG 73
G GG
Sbjct: 319 GYNGG 333
Score = 31.6 bits (70), Expect = 0.047
Identities = 22/65 (33%), Positives = 26/65 (39%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG + G S + G G + GGG GGGG GGG+G GGG
Sbjct: 484 GGSYNHGGSYNHGGGSYNHGGGSY---------------HHGGGGYNHGGGGHGGHGGGG 618
Query: 69 GGCGG 73
G G
Sbjct: 619 HGGHG 633
Score = 29.6 bits (65), Expect(2) = 0.001
Identities = 20/44 (45%), Positives = 23/44 (51%), Gaps = 6/44 (13%)
Frame = +1
Query: 33 VIEKVVMVEGDCSGGDSGGG---GGGGDGGGNGCGSG---GGGG 70
V+EK V GG + GG GGG +GGG G G GGGG
Sbjct: 148 VVEKTNEVNDAKYGGYNHGGYNHGGGYNGGGYNHGGGYHNGGGG 279
Score = 26.9 bits (58), Expect = 1.1
Identities = 12/27 (44%), Positives = 16/27 (58%)
Frame = -3
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVV 110
VVV T+TVV T V + +V VV+
Sbjct: 288 VVVTTTTVVVTTTVVITSTIVTTTVVI 208
Score = 26.6 bits (57), Expect(2) = 6e-05
Identities = 7/9 (77%), Positives = 7/9 (77%)
Frame = +3
Query: 74 LMWWRRRWW 82
L WWRR WW
Sbjct: 357 LQWWRRSWW 383
Score = 26.6 bits (57), Expect(2) = 0.001
Identities = 7/9 (77%), Positives = 7/9 (77%)
Frame = +3
Query: 74 LMWWRRRWW 82
L WWRR WW
Sbjct: 384 LQWWRRSWW 410
Score = 26.2 bits (56), Expect = 2.0
Identities = 12/25 (48%), Positives = 16/25 (64%)
Frame = -3
Query: 85 VVVTSTVVATAVVTVVVVVVVEAVV 109
VVVT+TVV T+ + VV+ VV
Sbjct: 267 VVVTTTVVITSTIVTTTVVITTMVV 193
Score = 26.2 bits (56), Expect = 2.0
Identities = 14/27 (51%), Positives = 17/27 (62%)
Frame = +2
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVV 110
VVVVT+TV + VV +VVEA V
Sbjct: 269 VVVVTTTVEVVTTMVEVVTMVVEATEV 349
Score = 25.4 bits (54), Expect = 3.3
Identities = 13/26 (50%), Positives = 16/26 (61%)
Frame = +2
Query: 85 VVVTSTVVATAVVTVVVVVVVEAVVV 110
+VV +TVV T V + VVV VVV
Sbjct: 200 MVVITTVVVTMVEVITTVVVTTTVVV 277
Score = 25.4 bits (54), Expect = 3.3
Identities = 16/30 (53%), Positives = 18/30 (59%), Gaps = 3/30 (10%)
Frame = -3
Query: 84 VVVVTSTVVATA---VVTVVVVVVVEAVVV 110
VVV TSTVV T VVT +VV +VV
Sbjct: 582 VVVTTSTVVVTTSSMVVTTSTMVVTSTMVV 493
Score = 25.4 bits (54), Expect = 3.3
Identities = 14/27 (51%), Positives = 15/27 (54%)
Frame = +2
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVV 110
VVV V+ T VVT VVVV V V
Sbjct: 218 VVVTMVEVITTVVVTTTVVVVTTTVEV 298
Score = 24.6 bits (52), Expect = 5.7
Identities = 12/26 (46%), Positives = 15/26 (57%)
Frame = +2
Query: 85 VVVTSTVVATAVVTVVVVVVVEAVVV 110
VVVT+TVV VV +VE V +
Sbjct: 251 VVVTTTVVVVTTTVEVVTTMVEVVTM 328
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 45.4 bits (106), Expect = 3e-06
Identities = 30/72 (41%), Positives = 36/72 (49%), Gaps = 7/72 (9%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGG----DSGGGG---GGGDGGGN 61
GG+ GG ++G GG + GGGG G + G + GGGG GGG GG
Sbjct: 247 GGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHG 426
Query: 62 GCGSGGGGGCGG 73
G G GGGG GG
Sbjct: 427 GGGYNGGGGHGG 462
Score = 45.4 bits (106), Expect(2) = 3e-07
Identities = 26/64 (40%), Positives = 30/64 (46%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG+ GG + G GG + GGGG +GGGG GG GGG G GG
Sbjct: 325 GGYNHGGGGYNHGGGGYNHGGGGY----------------NGGGGHGGHGGGGYNGGGGH 456
Query: 69 GGCG 72
GG G
Sbjct: 457 GGHG 468
Score = 41.6 bits (96), Expect = 5e-05
Identities = 34/90 (37%), Positives = 37/90 (40%), Gaps = 25/90 (27%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGG----GVVIEKVVMVE-------------GDCSGGDS-- 49
GG+ GG G G GG + GGG G V E G GG S
Sbjct: 388 GGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYN 567
Query: 50 --GG----GGGGGDGGGNGCGSGGGGGCGG 73
GG GGGG + GG G G GGGG GG
Sbjct: 568 HGGGSYHHGGGGYNHGGGGHGGHGGGGHGG 657
Score = 41.2 bits (95), Expect(2) = 2e-05
Identities = 26/67 (38%), Positives = 31/67 (45%), Gaps = 2/67 (2%)
Frame = +1
Query: 9 GGFADGGSSGDDGC--GGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSG 66
GG+ G + G G GG + GGGG + GG GGGG + GG G G
Sbjct: 157 GGYGGGYNHGGGGYNGGGYNHGGGGY----------NNGGGGYNHGGGGYNNGGGGYNHG 306
Query: 67 GGGGCGG 73
GGG GG
Sbjct: 307 GGGYNGG 327
Score = 33.1 bits (74), Expect(2) = 8e-04
Identities = 20/44 (45%), Positives = 22/44 (49%), Gaps = 3/44 (6%)
Frame = +1
Query: 33 VIEKVVMVEGDCSGGDSGG---GGGGGDGGGNGCGSGGGGGCGG 73
V+EK V GG GG GGGG +GGG G GG GG
Sbjct: 118 VVEKTNEVNDAKYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGG 249
Score = 25.0 bits (53), Expect = 4.4
Identities = 13/27 (48%), Positives = 16/27 (59%)
Frame = +2
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVV 110
VVV+ V+ TAVV V VV +VV
Sbjct: 188 VVVIMVEVITTAVVVTTTVEVVTTMVV 268
Score = 23.5 bits (49), Expect(2) = 8e-04
Identities = 15/29 (51%), Positives = 17/29 (57%), Gaps = 2/29 (6%)
Frame = +2
Query: 85 VVVTST--VVATAVVTVVVVVVVEAVVVE 111
VVVT+T VV T VV V V V +VE
Sbjct: 224 VVVTTTVEVVTTMVVVVTTTVEVATTMVE 310
Score = 22.7 bits (47), Expect(2) = 3e-07
Identities = 5/10 (50%), Positives = 7/10 (70%)
Frame = +3
Query: 74 LMWWRRRWWR 83
+ WW + WWR
Sbjct: 528 IWWWLQPWWR 557
Score = 21.2 bits (43), Expect(2) = 2e-05
Identities = 6/9 (66%), Positives = 6/9 (66%)
Frame = +3
Query: 74 LMWWRRRWW 82
L WWRR W
Sbjct: 435 LQWWRRSRW 461
>TC76563 homologue to PIR|C29356|C29356 hydroxyproline-rich glycoprotein
(clone Hyp2.13) - kidney bean (fragment), partial (34%)
Length = 507
Score = 48.5 bits (114), Expect = 4e-07
Identities = 32/68 (47%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDS--GGGGGGGDGGGNG 62
GG DG G D G GGD +GGGG + V+ GD GGD GGGG G+GGG
Sbjct: 333 GGGGDGEGGGGDL**YGGGGDGEGGGGDL***GVLGIGDGGGGDL**YGGGGDGEGGGGD 154
Query: 63 CGSGGGGG 70
GGGG
Sbjct: 153 L**YGGGG 130
Score = 45.1 bits (105), Expect = 4e-06
Identities = 34/73 (46%), Positives = 36/73 (48%), Gaps = 13/73 (17%)
Frame = -1
Query: 10 GFADGGSSGD---DGCGGDSDGGGGVVIEKVVMVEGDCSGGD----SGGGGGGGDG---- 58
G DGG GD G GGD +GGGG + EGD GGD GGG G G G
Sbjct: 468 GIGDGGG-GDL**YGGGGDGEGGGGDL***GGGGEGDGGGGDL**YGGGGDGEGGGGDL* 292
Query: 59 --GGNGCGSGGGG 69
GG G G GGGG
Sbjct: 291 *YGGGGDGEGGGG 253
Score = 41.2 bits (95), Expect = 6e-05
Identities = 34/91 (37%), Positives = 38/91 (41%), Gaps = 30/91 (32%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGD--------SGGGGGG- 55
GG DG G D G GG+ DGGGG + +G+ GGD G GGGG
Sbjct: 429 GGGGDGEGGGGDL***GGGGEGDGGGGDL**YGGGGDGEGGGGDL**YGGGGDGEGGGGD 250
Query: 56 ---------GDGG--------GNGCGSGGGG 69
GDGG G G G GGGG
Sbjct: 249 L***GVLGIGDGGGGDL**YGGGGDGEGGGG 157
Score = 35.0 bits (79), Expect(2) = 8e-04
Identities = 24/52 (46%), Positives = 27/52 (51%), Gaps = 8/52 (15%)
Frame = -1
Query: 27 DGGGGVVIEKVVMVEGDCSGGDS--GGGGGGGDGGG------NGCGSGGGGG 70
+GGGG + V GD GGD GGGG G+GGG G G G GGG
Sbjct: 507 EGGGGDL***GVWGIGDGGGGDL**YGGGGDGEGGGGDL***GGGGEGDGGG 352
Score = 21.6 bits (44), Expect(2) = 8e-04
Identities = 8/16 (50%), Positives = 10/16 (62%)
Frame = -3
Query: 68 GGGCGGLMWWRRRWWR 83
G G ++WW RRW R
Sbjct: 310 GWGRFVVVWWWRRW*R 263
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.145 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,871,173
Number of Sequences: 36976
Number of extensions: 91518
Number of successful extensions: 9493
Number of sequences better than 10.0: 1010
Number of HSP's better than 10.0 without gapping: 2504
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5240
length of query: 117
length of database: 9,014,727
effective HSP length: 93
effective length of query: 24
effective length of database: 5,575,959
effective search space: 133823016
effective search space used: 133823016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 50 (23.9 bits)
Lotus: description of TM0072a.11


