

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
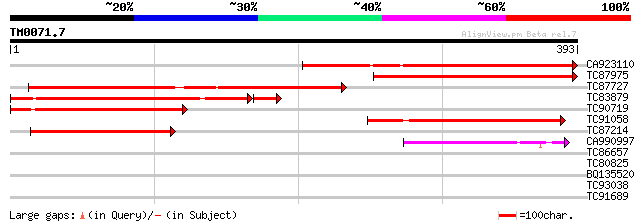
Query= TM0071.7
(393 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA923110 similar to GP|17529326|gb unknown protein {Arabidopsis ... 270 7e-73
TC87975 similar to GP|17529326|gb|AAL38890.1 unknown protein {Ar... 262 1e-70
TC87727 similar to PIR|T52392|T52392 hypothetical protein MMB12.... 220 6e-58
TC83879 similar to GP|17529326|gb|AAL38890.1 unknown protein {Ar... 201 1e-56
TC90719 similar to GP|17529326|gb|AAL38890.1 unknown protein {Ar... 214 6e-56
TC91058 similar to PIR|T52392|T52392 hypothetical protein MMB12.... 116 2e-26
TC87214 similar to GP|15451154|gb|AAK96848.1 Unknown protein {Ar... 111 5e-25
CA990997 similar to GP|12007447|gb magnesium transporter protein... 61 7e-10
TC86657 similar to GP|7108521|gb|AAF36454.1| ubiquitin-protein l... 33 0.15
TC80825 similar to PIR|F84614|F84614 probable kinesin heavy chai... 28 4.7
BQ135520 weakly similar to GP|7573600|dbj| ABC transporter homol... 28 4.7
TC93038 28 8.0
TC91689 similar to PIR|T48150|T48150 stress-induced protein sti1... 28 8.0
>CA923110 similar to GP|17529326|gb unknown protein {Arabidopsis thaliana},
partial (36%)
Length = 799
Score = 270 bits (690), Expect = 7e-73
Identities = 142/191 (74%), Positives = 159/191 (82%), Gaps = 1/191 (0%)
Frame = -1
Query: 204 LTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSSPVSGSGAANWFGGSPTIGSKISRA 262
LT RV K+R+ELE LLDDDDDMA+LYLSRK A SSSP S S NW P GSKI R+
Sbjct: 799 LTNRVHKIREELENLLDDDDDMAELYLSRKLAVSSSPSSSSDGPNWHY-IPYQGSKIHRS 623
Query: 263 SRASLATVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYIDDTEDYINIQLDNHRNQ 322
SR S AT++ ENDVEELEMLLEAYF+QIDGT NKLTT+REYIDDTEDYINIQLDNHRNQ
Sbjct: 622 SRGSAATLQ-GENDVEELEMLLEAYFIQIDGTLNKLTTVREYIDDTEDYINIQLDNHRNQ 446
Query: 323 LIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKWVVIVSGVFSAIMFLLII 382
LIQLELFLSSGTVCLS YSLVAAIFGMNIPY+W + H ++FKWVVI++G+ FL II
Sbjct: 445 LIQLELFLSSGTVCLSIYSLVAAIFGMNIPYTWKEGHAYVFKWVVILTGMVCGSFFLSII 266
Query: 383 VYARKKGLVGS 393
+YAR+KGLVGS
Sbjct: 265 LYARRKGLVGS 233
>TC87975 similar to GP|17529326|gb|AAL38890.1 unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 889
Score = 262 bits (670), Expect = 1e-70
Identities = 127/141 (90%), Positives = 136/141 (96%)
Frame = +2
Query: 253 PTIGSKISRASRASLATVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYIDDTEDYI 312
PT+GS+ISRASRAS+ TVR DENDVEELEMLLEAYFMQIDGT NKLTTLREYIDDTEDYI
Sbjct: 2 PTVGSRISRASRASIVTVRLDENDVEELEMLLEAYFMQIDGTLNKLTTLREYIDDTEDYI 181
Query: 313 NIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKWVVIVSGV 372
NIQLDNHRNQLIQLELFLSSGTVCL+FYSLVA IFGMNIPY+WND+HG+MFKWVVI SGV
Sbjct: 182 NIQLDNHRNQLIQLELFLSSGTVCLAFYSLVAGIFGMNIPYTWNDDHGYMFKWVVIFSGV 361
Query: 373 FSAIMFLLIIVYARKKGLVGS 393
FSAIMFL+II+YARKKGLVGS
Sbjct: 362 FSAIMFLMIIIYARKKGLVGS 424
>TC87727 similar to PIR|T52392|T52392 hypothetical protein MMB12.11
[imported] - Arabidopsis thaliana, partial (36%)
Length = 859
Score = 220 bits (561), Expect = 6e-58
Identities = 118/220 (53%), Positives = 152/220 (68%)
Frame = +1
Query: 14 AVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILG 73
AV ++KK S R W++ D G+ +++ K+AIM R + ARDLRILDPLLSYPST+LG
Sbjct: 109 AVPAIRKKGTSVRQWLVVDGTGEAQVIEAGKHAIMRRTGLPARDLRILDPLLSYPSTVLG 288
Query: 74 REKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVS 133
RE+AIV+NLEHIKAIITA EVLL + D +V P V+ELQ R+ L +
Sbjct: 289 RERAIVINLEHIKAIITANEVLLLNSRDPSVTPFVQELQARI-----LRHHEATTTPLPD 453
Query: 134 SQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLDR 193
+Q D+ + PFEF ALE LEA CS L + LE A+PALD+LTSKIS+ NL+R
Sbjct: 454 NQEDSHGGIK-ILPFEFVALEACLEAACSVLESEAKTLEQEAHPALDKLTSKISTLNLER 630
Query: 194 VRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK 233
VR++KS + +T RVQKVRDELE LLDDD+DMA++YL+ K
Sbjct: 631 VRQIKSRLVAITGRVQKVRDELEHLLDDDEDMAEMYLTEK 750
>TC83879 similar to GP|17529326|gb|AAL38890.1 unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 715
Score = 201 bits (511), Expect(2) = 1e-56
Identities = 116/169 (68%), Positives = 130/169 (76%), Gaps = 1/169 (0%)
Frame = +3
Query: 1 MARDGSVLPADPQAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRI 60
MA GS + QA+ KKKT SRSWIL D DG+ +LDVDKYAIM V+IHARDLRI
Sbjct: 162 MALAGSEVELKVQAMN--KKKTTISRSWILLDRDGRDIVLDVDKYAIMRLVEIHARDLRI 335
Query: 61 LDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLP-KVS 119
+DPLLSYPSTILGREK IVLNLEHIKAIITAEEVL+RDP D++VVPVVEEL+RRLP KVS
Sbjct: 336 MDPLLSYPSTILGREKVIVLNLEHIKAIITAEEVLVRDPMDDDVVPVVEELRRRLPLKVS 515
Query: 120 SLHQQQGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAART 168
Q Q + + V + E EE+E PFEFRALEV LEAICSFL ART
Sbjct: 516 VAGQGQVEEELCV---QEGEGGEENEFPFEFRALEVVLEAICSFLDART 653
Score = 37.0 bits (84), Expect(2) = 1e-56
Identities = 18/19 (94%), Positives = 18/19 (94%)
Frame = +1
Query: 170 ELEMAAYPALDELTSKISS 188
ELE AAYPALDELTSKISS
Sbjct: 658 ELETAAYPALDELTSKISS 714
>TC90719 similar to GP|17529326|gb|AAL38890.1 unknown protein {Arabidopsis
thaliana}, partial (26%)
Length = 704
Score = 214 bits (544), Expect = 6e-56
Identities = 110/123 (89%), Positives = 117/123 (94%)
Frame = +2
Query: 1 MARDGSVLPADPQAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRI 60
MARDGSV+PADPQA+ VKKKTQSSRSWI FD GQGSLLDVDKYAIMHRVQI+ARDLRI
Sbjct: 341 MARDGSVVPADPQAL--VKKKTQSSRSWIAFDGTGQGSLLDVDKYAIMHRVQINARDLRI 514
Query: 61 LDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSS 120
LDPLLSYPSTILGREKAIVLNLEHIKAIITA+EVLLRDPTDE+VVPVVEELQRRLPK+S
Sbjct: 515 LDPLLSYPSTILGREKAIVLNLEHIKAIITADEVLLRDPTDEHVVPVVEELQRRLPKLSD 694
Query: 121 LHQ 123
+HQ
Sbjct: 695 IHQ 703
>TC91058 similar to PIR|T52392|T52392 hypothetical protein MMB12.11
[imported] - Arabidopsis thaliana, partial (22%)
Length = 783
Score = 116 bits (290), Expect = 2e-26
Identities = 67/139 (48%), Positives = 94/139 (67%), Gaps = 2/139 (1%)
Frame = +3
Query: 249 FGGSPTIGSKISRASRA-SLATVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYIDD 307
FG S S+ +RAS S+ T + D VEELEMLLEAYF+QIDGT NKL+TLREY+DD
Sbjct: 111 FGASNLRDSRGTRASTTYSVTTTKLD---VEELEMLLEAYFVQIDGTLNKLSTLREYVDD 281
Query: 308 TEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGF-MFKWV 366
TEDYINI LD+ +N L+Q+ + L++ T+ +S + +VA IFGMNI D + + M +++
Sbjct: 282 TEDYINIMLDDKQNHLLQMGVMLTTATLVVSAFVVVAGIFGMNINIELFDKNLYGMREFM 461
Query: 367 VIVSGVFSAIMFLLIIVYA 385
V G + +FL ++ A
Sbjct: 462 WTVGGGTAGTIFLYVVAIA 518
>TC87214 similar to GP|15451154|gb|AAK96848.1 Unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 662
Score = 111 bits (277), Expect = 5e-25
Identities = 56/101 (55%), Positives = 73/101 (71%)
Frame = +2
Query: 15 VAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGR 74
V VKK+ Q +SWI D G +++VDK+ +M R + ARDLR+LDP+ YPSTILGR
Sbjct: 263 VVEVKKRGQGLKSWIRVDTSGNSQVIEVDKFTMMRRCDLPARDLRLLDPVFVYPSTILGR 442
Query: 75 EKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRL 115
EKAIV+NLE I+ IITA+EVLL + D+ V+ V +LQRRL
Sbjct: 443 EKAIVVNLEQIRCIITADEVLLLNSLDKYVLQYVIDLQRRL 565
>CA990997 similar to GP|12007447|gb magnesium transporter protein
{Arabidopsis thaliana}, partial (34%)
Length = 487
Score = 61.2 bits (147), Expect = 7e-10
Identities = 37/118 (31%), Positives = 65/118 (54%), Gaps = 3/118 (2%)
Frame = +2
Query: 274 ENDVEELEMLLEAYFMQIDGTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSG 333
E + EE+EMLLE Y + + + L + + ED I + L + R ++ ++EL L G
Sbjct: 122 EEEEEEIEMLLENYLQRCESCHGQAERLLDSAREMEDSIAVSLSSRRLEVSRVELLLQVG 301
Query: 334 TVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKWV---VIVSGVFSAIMFLLIIVYARKK 388
T C++ +LVA IFGMN+ S+ + F F + +I+ G+ ++F ++ Y R +
Sbjct: 302 TFCVAVGALVAGIFGMNLK-SYLEERVFAFWFTTSGIIIGGI---VVFFIMYNYLRAR 463
>TC86657 similar to GP|7108521|gb|AAF36454.1| ubiquitin-protein ligase 1
{Arabidopsis thaliana}, partial (6%)
Length = 1302
Score = 33.5 bits (75), Expect = 0.15
Identities = 54/232 (23%), Positives = 85/232 (36%), Gaps = 1/232 (0%)
Frame = +3
Query: 83 EHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHDTEAAE 142
E IKA+++ T N ++ LQ V+S +++ DG +S H
Sbjct: 183 ETIKALLS---------TSTNGAAILRVLQALSSFVNSSTEKENDG---ISRAH------ 308
Query: 143 EDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLDRVRKLKSAMT 202
FEF + ALE + L+ +++E + PA + +T SS V K S M
Sbjct: 309 -----FEFSEINSALEPLWHELSCCISKIESYSEPASEIITPSTSS-----VSKPSSVMP 458
Query: 203 RLTARVQKVRDELEQLLDDDDDMADLYLSRKAGSSSP-VSGSGAANWFGGSPTIGSKISR 261
L A Q ++ +E + + L + P +S A+ G +
Sbjct: 459 PLPAGSQNIQPYIESFFVVCEKLHPAQLGANNDNGVPYISDVEDASTSGTQQKTSGAAMK 638
Query: 262 ASRASLATVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYIDDTEDYIN 313
A V+F E LL A+ Q G K L I D+ N
Sbjct: 639 IDEKHGAFVKFSEKH----RKLLNAFIRQNPGLLEKSFALMLKIPRFIDFDN 782
>TC80825 similar to PIR|F84614|F84614 probable kinesin heavy chain
[imported] - Arabidopsis thaliana, partial (16%)
Length = 1771
Score = 28.5 bits (62), Expect = 4.7
Identities = 29/153 (18%), Positives = 63/153 (40%), Gaps = 5/153 (3%)
Frame = +2
Query: 71 ILGREKAIVLNLEHIKAIITAEEVLLRDPTDE--NVVPVVEELQRRLPKVSSLHQQQGDG 128
+L R K I N + K +T EV + N ++ ++ + ++H+ +
Sbjct: 638 VLLRSKLIQANFKRQKQCLTKHEVSAEVKKNP*GN*KKSLQNIESKAKGKDNIHKNLQEK 817
Query: 129 QEYVSSQHDTEAAEEDESPFEFRALEVAL---EAICSFLAARTTELEMAAYPALDELTSK 185
+ + Q + + + +++S + L L E C L + ELE L T+
Sbjct: 818 IKELEGQIELKTSMQNQSEKQVSQLCEKLKGKEETCCTLQHKVKELERKIKEQLQTETAN 997
Query: 186 ISSRNLDRVRKLKSAMTRLTARVQKVRDELEQL 218
+ D +KLK + + ++D++++L
Sbjct: 998 FQQKAWDLEKKLKDQLQGSESESSFLKDKIKEL 1096
>BQ135520 weakly similar to GP|7573600|dbj| ABC transporter homolog {Populus
nigra}, partial (17%)
Length = 747
Score = 28.5 bits (62), Expect = 4.7
Identities = 11/27 (40%), Positives = 18/27 (65%), Gaps = 1/27 (3%)
Frame = +2
Query: 340 YSLVAAIFGMNIP-YSWNDNHGFMFKW 365
Y +A++ M+IP ++WNDN +KW
Sbjct: 29 YLHLASMINMSIPAWTWNDNQMIQYKW 109
>TC93038
Length = 784
Score = 27.7 bits (60), Expect = 8.0
Identities = 24/87 (27%), Positives = 39/87 (44%), Gaps = 3/87 (3%)
Frame = -3
Query: 230 LSRKAGSSSPVSGSGAANWFGGSPTIGSKISRASRASLATVRFDENDVEELEMLL---EA 286
L + S++ + G+A F + T S + +A L V +ENDV E +
Sbjct: 542 LGERLSSTAKIYSPGSAE-FETASTRWSNL-QAPTVKLVVVPGNENDVAETVRFANEKDV 369
Query: 287 YFMQIDGTFNKLTTLREYIDDTEDYIN 313
F+ +G +TTL + D E Y+N
Sbjct: 368 PFLAYNGVHGAITTLGKMQDGIEIYLN 288
>TC91689 similar to PIR|T48150|T48150 stress-induced protein sti1-like
protein - Arabidopsis thaliana, partial (26%)
Length = 638
Score = 27.7 bits (60), Expect = 8.0
Identities = 16/60 (26%), Positives = 29/60 (47%)
Frame = +3
Query: 60 ILDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVS 119
+L + P ILGR E+ KA+ T +E L DP ++ ++ V +++ + S
Sbjct: 96 VLSLIRPSPRVILGRVXVQFFMKEYEKALETYQEGLKHDPNNQELLEGVRSCVKQINRTS 275
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,105,543
Number of Sequences: 36976
Number of extensions: 125210
Number of successful extensions: 671
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 664
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 666
length of query: 393
length of database: 9,014,727
effective HSP length: 98
effective length of query: 295
effective length of database: 5,391,079
effective search space: 1590368305
effective search space used: 1590368305
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0071.7


