

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0069.6
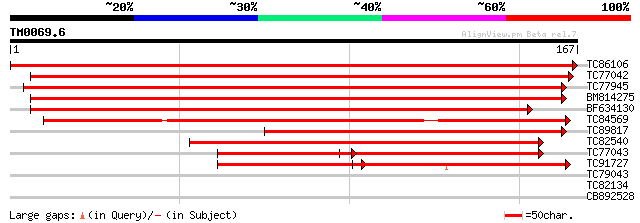
(167 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86106 homologue to GP|21068666|emb|CAD31839. putative phosphol... 302 4e-83
TC77042 similar to GP|14532478|gb|AAK63967.1 AT4g11600/T5C23_30 ... 252 4e-68
TC77945 similar to SP|O24296|GSHY_PEA Phospholipid hydroperoxide... 233 3e-62
BM814275 similar to GP|21592603|gb unknown {Arabidopsis thaliana... 233 3e-62
BF634130 similar to GP|21592603|gb unknown {Arabidopsis thaliana... 217 2e-57
TC84569 weakly similar to GP|727367|gb|AAA64283.1|| Hyr1p {Sacch... 139 7e-34
TC89817 similar to GP|6179604|emb|CAB59895.1 glutathione peroxid... 120 3e-28
TC82540 similar to PIR|H83292|H83292 probable glutathione peroxi... 119 6e-28
TC77043 similar to PIR|S33618|S33618 glutathione peroxidase (EC ... 94 3e-20
TC91727 weakly similar to SP|Q9LYB4|GSHZ_ARATH Probable glutathi... 80 5e-16
TC79043 homologue to GP|6899842|dbj|BAA90524.1 peroxiredoxin Q {... 33 0.070
TC82134 similar to PIR|T07949|T07949 calcium binding protein - l... 27 3.8
CB892528 weakly similar to PIR|E96647|E966 hypothetical protein ... 26 6.5
>TC86106 homologue to GP|21068666|emb|CAD31839. putative phospholipid
hydroperoxide glutathione peroxidase {Cicer arietinum},
complete
Length = 893
Score = 302 bits (774), Expect = 4e-83
Identities = 142/167 (85%), Positives = 160/167 (95%)
Frame = +3
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS 60
MAE +SKS+YDFTVKDI GNDVSLSQY GKVL++VNVASQCGLTQTNYKELN+LY+KYK
Sbjct: 129 MAENSSKSIYDFTVKDISGNDVSLSQYRGKVLLVVNVASQCGLTQTNYKELNVLYQKYKD 308
Query: 61 KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK 120
+ EILAFPCNQF GQEPG+++EIQ+VVCTRFK+EFPVFDKVEVNGKNAEPL+KFLKDQK
Sbjct: 309 QDFEILAFPCNQFRGQEPGSSEEIQNVVCTRFKAEFPVFDKVEVNGKNAEPLYKFLKDQK 488
Query: 121 GGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQSS 167
GGIFGDGIKWNFTKFLVNKEGKVV+RYAPTT+P+KIEKD+EKLL+SS
Sbjct: 489 GGIFGDGIKWNFTKFLVNKEGKVVDRYAPTTAPLKIEKDIEKLLRSS 629
>TC77042 similar to GP|14532478|gb|AAK63967.1 AT4g11600/T5C23_30
{Arabidopsis thaliana}, partial (74%)
Length = 1015
Score = 252 bits (644), Expect = 4e-68
Identities = 118/160 (73%), Positives = 139/160 (86%)
Frame = +1
Query: 7 KSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEIL 66
+S++DFTVKD +GNDV+L Y GKVLIIVNVASQCGLT +NY EL+ LYEKYKSKGLEIL
Sbjct: 259 QSIHDFTVKDAKGNDVNLGDYKGKVLIIVNVASQCGLTNSNYTELSQLYEKYKSKGLEIL 438
Query: 67 AFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGD 126
AFPCNQF QEPG+ +EIQ+ VCTRFK+EFPVFDKV+VNG A P++K+LK KGG+FGD
Sbjct: 439 AFPCNQFGAQEPGSVEEIQNFVCTRFKAEFPVFDKVDVNGATAAPIYKYLKSSKGGLFGD 618
Query: 127 GIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQS 166
GIKWNF+KFLV+K G VV+RYAPTTSP+ IEKDL KLL +
Sbjct: 619 GIKWNFSKFLVDKNGNVVDRYAPTTSPLSIEKDLLKLLDA 738
>TC77945 similar to SP|O24296|GSHY_PEA Phospholipid hydroperoxide
glutathione peroxidase chloroplast precursor (EC
1.11.1.9) (PHGPx)., complete
Length = 1006
Score = 233 bits (594), Expect = 3e-62
Identities = 108/160 (67%), Positives = 131/160 (81%)
Frame = +1
Query: 5 TSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLE 64
T KS+YDFTVKDI DV LS++ GKVL+IVNVAS+CGLT +NY EL+ LYE +K KGLE
Sbjct: 310 TDKSIYDFTVKDIDKKDVPLSKFKGKVLLIVNVASRCGLTSSNYTELSHLYENFKDKGLE 489
Query: 65 ILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIF 124
+LAFPCNQF QEPG+N+EI+ CTRFK+EFP+FDKV+VNG P+++FLK GG F
Sbjct: 490 VLAFPCNQFGMQEPGSNEEIKKFACTRFKAEFPIFDKVDVNGPFTAPVYQFLKSSSGGFF 669
Query: 125 GDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
GD +KWNF KFLV+K GKVVERY PTTSP +IEKD++KLL
Sbjct: 670 GDLVKWNFEKFLVDKNGKVVERYPPTTSPFQIEKDIQKLL 789
>BM814275 similar to GP|21592603|gb unknown {Arabidopsis thaliana}, partial
(96%)
Length = 699
Score = 233 bits (594), Expect = 3e-62
Identities = 109/158 (68%), Positives = 132/158 (82%)
Frame = +2
Query: 7 KSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEIL 66
KS+YDFT+KD GNDV L+ Y GKVL+IVNVAS+CG+T +NY LN LY+KYK KGLEIL
Sbjct: 71 KSVYDFTLKDGMGNDVDLATYKGKVLLIVNVASKCGMTNSNYVGLNQLYDKYKLKGLEIL 250
Query: 67 AFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGD 126
AFP NQF +EPGTND+I D VCT FKSEFP+FDK+EVNG N+ PL+KFLK K GIFGD
Sbjct: 251 AFPSNQFGEEEPGTNDQILDFVCTHFKSEFPIFDKIEVNGDNSAPLYKFLKSGKWGIFGD 430
Query: 127 GIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
I+WNF KFLV+K+G+VV+RY PTTSP+ +E+D+ KLL
Sbjct: 431 DIQWNFAKFLVDKDGQVVDRYYPTTSPLSLERDICKLL 544
>BF634130 similar to GP|21592603|gb unknown {Arabidopsis thaliana}, partial
(90%)
Length = 563
Score = 217 bits (552), Expect = 2e-57
Identities = 102/148 (68%), Positives = 120/148 (80%)
Frame = +2
Query: 7 KSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEIL 66
KS+YDFT+KD GNDV L+ Y GKVL+IVNVAS+CG+T +NY LN LY+KYK KGLEIL
Sbjct: 50 KSVYDFTLKDGMGNDVDLATYKGKVLLIVNVASKCGMTNSNYVGLNQLYDKYKLKGLEIL 229
Query: 67 AFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGD 126
AFP NQF +EPGTND+I D VCT FKS FP+FDK+EVNG N+ PL+KFLK K GIFGD
Sbjct: 230 AFPSNQFGEEEPGTNDQILDFVCTHFKSXFPIFDKIEVNGDNSAPLYKFLKSGKWGIFGD 409
Query: 127 GIKWNFTKFLVNKEGKVVERYAPTTSPM 154
I WNF KFLV +G+VV+RY PTTSP+
Sbjct: 410 DIXWNFAKFLVXXDGQVVDRYYPTTSPL 493
>TC84569 weakly similar to GP|727367|gb|AAA64283.1|| Hyr1p {Saccharomyces
cerevisiae}, partial (79%)
Length = 564
Score = 139 bits (349), Expect = 7e-34
Identities = 67/155 (43%), Positives = 95/155 (61%)
Frame = +3
Query: 11 DFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEILAFPC 70
+F DI+ + S + GKV+++VNVAS+CG T Y L L+ KYK +GL + PC
Sbjct: 3 NFEALDIKKKPFNFSAWKGKVVLLVNVASKCGYTP-QYTGLESLHNKYKDQGLIVAGLPC 179
Query: 71 NQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGDGIKW 130
NQF QEPGT ++I + FP+F K++VNG+N P++K+LK Q+ G IKW
Sbjct: 180 NQFGAQEPGTEEDIVSFCSVTYNVTFPLFAKIDVNGENESPVYKYLKSQQPG----DIKW 347
Query: 131 NFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQ 165
NF KFL++K G VV+RY P I D+E L++
Sbjct: 348 NFEKFLIDKNGNVVKRYTSAAKPEDIASDIENLIK 452
>TC89817 similar to GP|6179604|emb|CAB59895.1 glutathione peroxidase-like
protein GPX54Hv {Hordeum vulgare}, partial (53%)
Length = 584
Score = 120 bits (300), Expect = 3e-28
Identities = 52/89 (58%), Positives = 68/89 (75%)
Frame = +2
Query: 76 QEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGDGIKWNFTKF 135
QEPG + E + CTRFK+E+P+F K+ VNG + PL+KFLK++K G G IKWNFTKF
Sbjct: 5 QEPGNSLEAEQFACTRFKAEYPIFGKIRVNGPDTAPLYKFLKEKKSGFLGSRIKWNFTKF 184
Query: 136 LVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
LV+KEG V++RY+PTTSP IE D++K L
Sbjct: 185 LVDKEGHVLQRYSPTTSPFSIENDIKKAL 271
>TC82540 similar to PIR|H83292|H83292 probable glutathione peroxidase PA2826
[imported] - Pseudomonas aeruginosa (strain PAO1),
partial (62%)
Length = 678
Score = 119 bits (298), Expect = 6e-28
Identities = 54/104 (51%), Positives = 72/104 (68%)
Frame = +2
Query: 54 LYEKYKSKGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLF 113
LY KY +GL +L FPCNQF QEPG DEI + + FP+F+KVEVNG A P+F
Sbjct: 8 LYTKYAGQGLVVLGFPCNQFGKQEPGGADEISNTCYINYGVSFPMFEKVEVNGGAAHPVF 187
Query: 114 KFLKDQKGGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIE 157
+FLK + G+ G IKWNFTKFL+ ++G ++R+AP T+P K+E
Sbjct: 188 RFLKKELPGVLGGRIKWNFTKFLIGRDGIPLKRFAPMTTPEKME 319
>TC77043 similar to PIR|S33618|S33618 glutathione peroxidase (EC 1.11.1.9) -
sweet orange, partial (57%)
Length = 1748
Score = 94.0 bits (232), Expect = 3e-20
Identities = 41/60 (68%), Positives = 51/60 (84%)
Frame = +3
Query: 98 VFDKVEVNGKNAEPLFKFLKDQKGGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIE 157
VF+ V+VNG A P++K+LK KGG+FGDGIKWNF+KFLV+K G VV+RYAPTTSP+ IE
Sbjct: 1014 VFN*VDVNGATAAPIYKYLKSSKGGLFGDGIKWNFSKFLVDKNGNVVDRYAPTTSPLSIE 1193
Score = 74.3 bits (181), Expect = 2e-14
Identities = 33/41 (80%), Positives = 37/41 (89%)
Frame = +1
Query: 62 GLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKV 102
GLEILAFPCNQF QEPG+ +EIQ+ VCTRFK+EFPVFDKV
Sbjct: 208 GLEILAFPCNQFGAQEPGSVEEIQNFVCTRFKAEFPVFDKV 330
>TC91727 weakly similar to SP|Q9LYB4|GSHZ_ARATH Probable glutathione
peroxidase At3g63080 (EC 1.11.1.9). [Mouse-ear cress]
{Arabidopsis thaliana}, partial (57%)
Length = 1124
Score = 79.7 bits (195), Expect = 5e-16
Identities = 35/65 (53%), Positives = 51/65 (77%), Gaps = 1/65 (1%)
Frame = +3
Query: 102 VEVNGKNAEPLFKFLKDQKGGIFGDG-IKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDL 160
+ VNG++ P++K+LK QK G G IKWNFTKFLV++EG+V++RY+PTT P+ IE D+
Sbjct: 624 IRVNGQDTAPVYKYLKSQKCGSLGSRRIKWNFTKFLVDEEGRVIQRYSPTTPPLAIENDI 803
Query: 161 EKLLQ 165
+K L+
Sbjct: 804 KKALR 818
Score = 61.2 bits (147), Expect = 2e-10
Identities = 26/44 (59%), Positives = 32/44 (72%)
Frame = +1
Query: 62 GLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVN 105
GLEIL FPCNQF +EPGT+ E QD C R+K+E+P+ KV N
Sbjct: 310 GLEILGFPCNQFLRKEPGTSQEAQDFACDRYKAEYPILGKV*YN 441
>TC79043 homologue to GP|6899842|dbj|BAA90524.1 peroxiredoxin Q {Sedum
lineare}, partial (81%)
Length = 935
Score = 32.7 bits (73), Expect = 0.070
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Frame = +3
Query: 12 FTVKDIRGNDVSLSQYSGKVLII----VNVASQCGLTQTNYKELNILYEKYKSKGLEIL 66
FT+KD G VSLS+Y GK +++ + C +++ YEK+K G E++
Sbjct: 273 FTLKDQDGKTVSLSKYKGKPVVVYFYPADETPGCTKQACAFRD---SYEKFKKAGAEVV 440
>TC82134 similar to PIR|T07949|T07949 calcium binding protein - loblolly
pine, partial (27%)
Length = 630
Score = 26.9 bits (58), Expect = 3.8
Identities = 12/23 (52%), Positives = 16/23 (69%), Gaps = 1/23 (4%)
Frame = +1
Query: 126 DGIKWNFTKFLVNKEGKV-VERY 147
D +KW F KF NK+GK+ +E Y
Sbjct: 64 DEMKWVFEKFDTNKDGKISLEEY 132
>CB892528 weakly similar to PIR|E96647|E966 hypothetical protein F19K23.5
[imported] - Arabidopsis thaliana, partial (7%)
Length = 549
Score = 26.2 bits (56), Expect = 6.5
Identities = 13/32 (40%), Positives = 22/32 (68%), Gaps = 1/32 (3%)
Frame = -1
Query: 134 KFLVNKEGKVVERYAPTTSPMKIEK-DLEKLL 164
KF +N KVVE +TSP+++E+ D+ ++L
Sbjct: 354 KFDINSAWKVVEIAMSSTSPIEVERPDMSQIL 259
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,460,119
Number of Sequences: 36976
Number of extensions: 53051
Number of successful extensions: 183
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 180
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 181
length of query: 167
length of database: 9,014,727
effective HSP length: 89
effective length of query: 78
effective length of database: 5,723,863
effective search space: 446461314
effective search space used: 446461314
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0069.6


