

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0067b.6
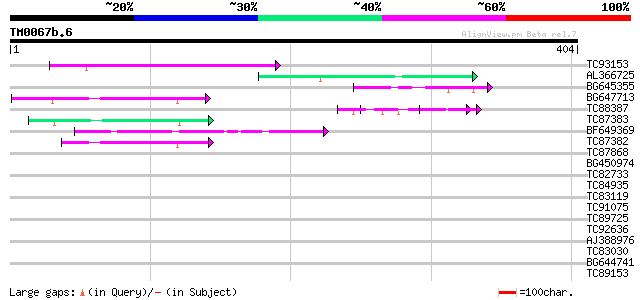
(404 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 107 6e-24
AL366725 59 4e-09
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 51 9e-07
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 46 2e-05
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 45 5e-05
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 43 2e-04
BF649369 43 2e-04
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 42 3e-04
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 38 0.006
BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR p... 36 0.023
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 36 0.030
TC84935 similar to PIR|G96631|G96631 probable RNA-binding protei... 36 0.030
TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 35 0.052
TC91075 similar to PIR|T48025|T48025 hypothetical protein T12C14... 35 0.068
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 35 0.068
TC92636 homologue to GP|15042313|gb|AAK82093.1 232R {Chilo iride... 34 0.089
AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing fact... 34 0.12
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 33 0.15
BG644741 32 0.34
TC89153 similar to GP|18855061|gb|AAL79753.1 putative RNA helica... 32 0.44
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 107 bits (268), Expect = 6e-24
Identities = 57/167 (34%), Positives = 90/167 (53%), Gaps = 2/167 (1%)
Frame = +2
Query: 29 ANDNAMRRATEEAREQHQRQREITL--DQNKGLNDFRRQNPPKFSGGTDPDKADLWIQEV 86
++D A+ A E + Q+ ++ D + L F R +PP F G PD A W++E+
Sbjct: 11 SSDAALVAALEAVAQAVQQLPKVDTGSDGTRMLETFLRNHPPTFKGRYAPDGA*KWLKEI 190
Query: 87 EKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEEISWNSFRTAFLEKYFPT 146
E+IF V+Q E KV+ T++L +A+ WW ++E + ++W FR FL +YFP
Sbjct: 191 ERIFRVMQCFETQKVQFGTHMLAEEADDWWISLLPVLEQDDAVVTWAMFRKEFLGRYFPE 370
Query: 147 SARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNERYMC 193
R ++E +FL L+QG MSV E+A+K LA + + E C
Sbjct: 371 DVRGKKEIEFLELKQGDMSVTEYAAKFVELATFYPHYSAETAEFSKC 511
>AL366725
Length = 485
Score = 58.5 bits (140), Expect = 4e-09
Identities = 42/164 (25%), Positives = 65/164 (39%), Gaps = 8/164 (4%)
Frame = +2
Query: 178 KHFQFFHDHVNERYMCKRFVNGLRPDIEDSVRPLRIMRFQSLV-------EKATEVELMK 230
K + + E C +F NGLRPDI+ ++ ++ F LV E + +
Sbjct: 2 KFYPHYAAETAEFSKCIKFENGLRPDIKRAIGYQQLRVFPDLVNTCRIYEEDTKAHDKVV 181
Query: 231 NRRLNRAGTGGPMRSGSQNFQSREKF-QSRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQT 289
N R + P + + +++ R P ++ A +Y G G +
Sbjct: 182 NERKTKGQ*SRPKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNY----GEKGHKSNVC 349
Query: 290 LKKETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQCRAPK 333
K+ C RC + GH C CFNCN GH QC+ PK
Sbjct: 350 PKEIKKCVRCDKKGHIVADCKRNDIVCFNCNEEGHIGSQCKQPK 481
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 50.8 bits (120), Expect = 9e-07
Identities = 30/116 (25%), Positives = 50/116 (42%), Gaps = 17/116 (14%)
Frame = -1
Query: 246 GSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHY 305
G + S + R P ++ AG + + ++G+ G SG C++C +PGH+
Sbjct: 615 GGSDHSSAQCLHLRNPPEQTAGGAYVN----TVSGSGGASGK--------CYKCQQPGHW 472
Query: 306 ANACPD-------------TRPKCFNCNRMGHTAGQC----RAPKTEPTVNTARGK 344
A+ CP C+ CN+ GH A C AP++ NT +G+
Sbjct: 471 ASNCPSMSAANRVSGGSGGASGNCYKCNQPGHWANNCPNMSAAPQSHGNSNTGQGR 304
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 46.2 bits (108), Expect = 2e-05
Identities = 32/147 (21%), Positives = 60/147 (40%), Gaps = 5/147 (3%)
Frame = +2
Query: 2 VNTNQLAEMMATMAQAVTAQAQAVTAQA---NDNAMRRATEEAREQHQRQREITLDQNKG 58
+ ++ + + + V AQ + AQ +D+ + +R +R+ + + K
Sbjct: 161 IEMEEMRRQIQLLQETVNAQQALLEAQRRRNDDDGSGSDSSSSRSSRSHRRQTRMSKIK- 337
Query: 59 LNDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKG 118
+ P F G PD+ W+Q +E++F + AE KV++ L A WWK
Sbjct: 338 ------VDIPDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKVKIVAAKLKKHASIWWKN 499
Query: 119 --TRGIMEDNHEEISWNSFRTAFLEKY 143
+ E + +W+ R KY
Sbjct: 500 LKRKRNCEGKSKIKTWDKMRQKLTRKY 580
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 45.1 bits (105), Expect = 5e-05
Identities = 17/44 (38%), Positives = 26/44 (58%)
Frame = +1
Query: 293 ETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQCRAPKTEP 336
++ C+ C EPGH A++CP+ C C + GH A +C P+ P
Sbjct: 529 KSLCWNCKEPGHMASSCPN-EGICHTCGKAGHRARECTVPQKPP 657
Score = 43.5 bits (101), Expect = 1e-04
Identities = 24/84 (28%), Positives = 33/84 (38%), Gaps = 5/84 (5%)
Frame = +1
Query: 251 QSREKFQSRGPYQR-----PAGTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHY 305
+SR K +SR P R P S + + ++ C C PGHY
Sbjct: 274 RSRSKSRSRSPMDRSRSRSPVDRRIRSERFSHREAPYRRDSRRGFSQDNLCKNCKRPGHY 453
Query: 306 ANACPDTRPKCFNCNRMGHTAGQC 329
CP+ C NC+ GH A +C
Sbjct: 454 VRECPNV-AVCHNCSLPGHIASEC 522
Score = 42.4 bits (98), Expect = 3e-04
Identities = 30/103 (29%), Positives = 44/103 (42%), Gaps = 7/103 (6%)
Frame = +1
Query: 234 LNRAGTGGPM--RSGSQNFQSREKFQSRGPYQRPAGTGFTSGSY-----RPMTGAAGGSG 286
++R+ + P+ R S+ F RE PY+R + GF+ + RP G
Sbjct: 307 MDRSRSRSPVDRRIRSERFSHRE-----APYRRDSRRGFSQDNLCKNCKRP------GHY 453
Query: 287 DQTLKKETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQC 329
+ C C PGH A+ C T+ C+NC GH A C
Sbjct: 454 VRECPNVAVCHNCSLPGHIASEC-STKSLCWNCKEPGHMASSC 579
Score = 37.0 bits (84), Expect = 0.014
Identities = 17/37 (45%), Positives = 19/37 (50%)
Frame = +1
Query: 293 ETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQC 329
E C C + GH A CP+ P C CN GH A QC
Sbjct: 721 EKACNNCRKTGHLARDCPND-PICNLCNISGHVARQC 828
Score = 34.3 bits (77), Expect = 0.089
Identities = 17/47 (36%), Positives = 20/47 (42%), Gaps = 6/47 (12%)
Frame = +1
Query: 293 ETTCFRCGEPGHYANAC------PDTRPKCFNCNRMGHTAGQCRAPK 333
E C CG+ GH A C P C NC + GH A +C K
Sbjct: 586 EGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVECTNEK 726
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 43.1 bits (100), Expect = 2e-04
Identities = 33/137 (24%), Positives = 54/137 (39%), Gaps = 5/137 (3%)
Frame = -2
Query: 14 MAQAVTAQAQAVTAQAN---DNAMRRATEEAREQHQRQREITLDQNKGLNDFRRQNPPKF 70
+ + V AQ + AQ D+ + +R ++RE ++ K + P F
Sbjct: 874 LQEIVNAQQALLEAQQKRFKDHVSSSDSLSSRSSRSQRREFQMNDIK*-------DIPDF 716
Query: 71 SGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGT--RGIMEDNHE 128
G PD W+Q +E++F + E KV++ L A WW+ R E +
Sbjct: 715 EGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRRRKREGKSK 536
Query: 129 EISWNSFRTAFLEKYFP 145
+W R KY P
Sbjct: 535 IKTWEKMRQKLTRKYLP 485
>BF649369
Length = 631
Score = 43.1 bits (100), Expect = 2e-04
Identities = 42/181 (23%), Positives = 74/181 (40%)
Frame = +3
Query: 47 RQREITLDQNKGLNDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATY 106
R+ + QN+ ++ P F G D WI E F V T + +V+++
Sbjct: 72 REEYVESSQNESRLAGKKVKLPLFEG----DDPVAWITRAEIYFDVQNTPDDMRVKLSRL 239
Query: 107 LLLGDAEYWWKGTRGIMEDNHEEISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSV 166
+ G +W+ ++ + +++S + A + +Y + E + TLRQ G SV
Sbjct: 240 SMEGPTIHWF----NLLMETEDDLSREKLKKALIARYDGRRLENPFE-ELSTLRQIG-SV 401
Query: 167 PEFASKLESLAKHFQFFHDHVNERYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEKATEV 226
EF E L+ + E F++GL+ I VR L ++ A +V
Sbjct: 402 EEFVEAFELLSSQV----GRLPEEQYLGYFMSGLKAHIRRRVRTLNPTTRMQMMRIAKDV 569
Query: 227 E 227
E
Sbjct: 570 E 572
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 42.4 bits (98), Expect = 3e-04
Identities = 26/110 (23%), Positives = 46/110 (41%), Gaps = 2/110 (1%)
Frame = +2
Query: 38 TEEAREQHQRQREITLDQNKGLNDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAE 97
+ +R H ++R++ ++ K + P F G D W+Q +E++F + E
Sbjct: 560 SSSSRSSHSQRRQLQMNDIK-------VDIPDFEGNLQLDDFLDWLQTIERVFEYKEVPE 718
Query: 98 GAKVRMATYLLLGDAEYWWKG--TRGIMEDNHEEISWNSFRTAFLEKYFP 145
KV++ L A WW+ R E + +W+ R KY P
Sbjct: 719 EQKVKIVAAKLKKHALIWWENLKRRRKREGKSKIKTWDKMRQKLTRKYLP 868
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 38.1 bits (87), Expect = 0.006
Identities = 24/68 (35%), Positives = 28/68 (40%)
Frame = +3
Query: 264 RPAGTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANACPDTRPKCFNCNRMG 323
R G G G R G GG D C+ CGEPGH+A C NR G
Sbjct: 258 RSGGGGGGGGGGRGRGGGGGGGSD------LKCYECGEPGHFARECR---------NRGG 392
Query: 324 HTAGQCRA 331
AG+ R+
Sbjct: 393 GGAGRRRS 416
>BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (54%)
Length = 364
Score = 36.2 bits (82), Expect = 0.023
Identities = 15/30 (50%), Positives = 17/30 (56%)
Frame = +1
Query: 280 GAAGGSGDQTLKKETTCFRCGEPGHYANAC 309
G GG G LK C+ CGEPGH+A C
Sbjct: 283 GGRGGRGGDDLK----CYECGEPGHFAREC 360
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 35.8 bits (81), Expect = 0.030
Identities = 18/50 (36%), Positives = 21/50 (42%), Gaps = 5/50 (10%)
Frame = +3
Query: 291 KKETTCFRCGEPGHYANACPDTRPK-----CFNCNRMGHTAGQCRAPKTE 335
+K C RC GH A CPD K +NC GH+ C P E
Sbjct: 342 EKNKICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQE 491
Score = 28.1 bits (61), Expect = 6.3
Identities = 12/40 (30%), Positives = 16/40 (40%), Gaps = 7/40 (17%)
Frame = +3
Query: 297 FRCGEPGHYANACPDTRPK-------CFNCNRMGHTAGQC 329
+ CG+ GH CP + CF C GH + C
Sbjct: 435 YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQGHLSKNC 554
>TC84935 similar to PIR|G96631|G96631 probable RNA-binding protein F8A5.17
[imported] - Arabidopsis thaliana, partial (41%)
Length = 552
Score = 35.8 bits (81), Expect = 0.030
Identities = 17/42 (40%), Positives = 25/42 (59%)
Frame = +2
Query: 269 GFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANACP 310
GF+SG + G+GD+ + + CF+CG PGH+A CP
Sbjct: 413 GFSSGGR-----GSYGAGDRVGQDD--CFKCGRPGHWARDCP 517
>TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 421
Score = 35.0 bits (79), Expect = 0.052
Identities = 29/95 (30%), Positives = 43/95 (44%)
Frame = +3
Query: 225 EVELMKNRRLNRAGTGGPMRSGSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAAGG 284
EV+ KN +++ RS S++ +SR + +SR P R + SYR
Sbjct: 165 EVKKEKNLKMS-----SDSRSRSRS-RSRSRSRSRSPRIRKIRSD--RHSYRDAPYRRDS 320
Query: 285 SGDQTLKKETTCFRCGEPGHYANACPDTRPKCFNC 319
S + ++ C C PGHYA CP+ C NC
Sbjct: 321 S--RGFSRDNLCKNCKRPGHYARECPNV-AVCHNC 416
>TC91075 similar to PIR|T48025|T48025 hypothetical protein T12C14.30 -
Arabidopsis thaliana, partial (29%)
Length = 686
Score = 34.7 bits (78), Expect = 0.068
Identities = 29/114 (25%), Positives = 45/114 (39%), Gaps = 3/114 (2%)
Frame = +2
Query: 202 PDIEDSVRPLRIMRFQSLVEKATEVELMKNRRLNRAGTGGPMRSGSQNFQSREKFQSRGP 261
PDI+D R L ++ A + + + N+ G S E+ + R P
Sbjct: 146 PDIDDDFRDL----YKEYTGPAGSGSVQERAKSNKRSNAG----------SDEEDEVRDP 283
Query: 262 YQRPAGTGFTSGS---YRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANACPDT 312
P TGF S + + A + + ++E C CGE GH+ CP T
Sbjct: 284 NAIP--TGFISRDAKVWEAKSKATERNWKKRKEEEMICKLCGESGHFTQGCPST 439
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 34.7 bits (78), Expect = 0.068
Identities = 17/45 (37%), Positives = 21/45 (45%)
Frame = +1
Query: 266 AGTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANACP 310
+G G G Y G GG G +C+ CGE GH+A CP
Sbjct: 61 SGGGGGGGRYGGGGGGGGGGGGGG-----SCYSCGESGHFARDCP 180
Score = 28.5 bits (62), Expect = 4.9
Identities = 14/54 (25%), Positives = 17/54 (30%), Gaps = 20/54 (37%)
Frame = +1
Query: 296 CFRCGEPGHYANACPD--------------------TRPKCFNCNRMGHTAGQC 329
C+ CGE GH A C C++C GH A C
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDC 177
>TC92636 homologue to GP|15042313|gb|AAK82093.1 232R {Chilo iridescent
virus}, partial (1%)
Length = 772
Score = 34.3 bits (77), Expect = 0.089
Identities = 25/101 (24%), Positives = 42/101 (40%), Gaps = 3/101 (2%)
Frame = +3
Query: 3 NTNQLAEM-MATMAQAVTAQAQAVTAQAN--DNAMRRATEEAREQHQRQREITLDQNKGL 59
N L EM M M + + + V AQ + RR ++ R + K L
Sbjct: 234 NARGLQEMEMEEMRRQIQELQETVNAQQAILEAERRRVDDDGSSDSSSSRSSRSHRRKTL 413
Query: 60 NDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAEGAK 100
+ + + P F G PD+ W+Q +E++F + GA+
Sbjct: 414 MNDIKVDIPDFEGELQPDEFVDWLQAIERVFEYKEIPRGAQ 536
>AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (62%)
Length = 508
Score = 33.9 bits (76), Expect = 0.12
Identities = 13/30 (43%), Positives = 17/30 (56%)
Frame = +2
Query: 280 GAAGGSGDQTLKKETTCFRCGEPGHYANAC 309
G +GG G + C+ CGEPGH+A C
Sbjct: 311 GRSGGGGS-----DLKCYXCGEPGHFARXC 385
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 33.5 bits (75), Expect = 0.15
Identities = 13/23 (56%), Positives = 15/23 (64%)
Frame = +1
Query: 295 TCFRCGEPGHYANACPDTRPKCF 317
TCF CGE GH A+ CP+ R F
Sbjct: 163 TCFTCGESGHRASDCPNKRGDFF 231
>BG644741
Length = 735
Score = 32.3 bits (72), Expect = 0.34
Identities = 22/84 (26%), Positives = 38/84 (45%), Gaps = 5/84 (5%)
Frame = -2
Query: 101 VRMATYLLLGDAEYWWKGTRGIMEDNHEEISWNSFRTAFLEKYFPTSAR---DERESQFL 157
+R+ L+G+A+ W+ + +WN R FL +Y+P S + ++R + F+
Sbjct: 587 LRVFPLSLMGEADIWFTEL-----PYNSIFTWNQLRDVFLARYYPVSKKLNHNDRVNNFV 423
Query: 158 TL--RQGGMSVPEFASKLESLAKH 179
L S F S L S+ H
Sbjct: 422 ALPGESVSSSWDRFTSFLRSVPNH 351
>TC89153 similar to GP|18855061|gb|AAL79753.1 putative RNA helicase {Oryza
sativa}, partial (3%)
Length = 737
Score = 32.0 bits (71), Expect = 0.44
Identities = 11/18 (61%), Positives = 14/18 (77%)
Frame = +3
Query: 296 CFRCGEPGHYANACPDTR 313
CF CG+PGH A+ CP+ R
Sbjct: 147 CFSCGQPGHRASDCPN*R 200
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,072,901
Number of Sequences: 36976
Number of extensions: 148095
Number of successful extensions: 836
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 791
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 818
length of query: 404
length of database: 9,014,727
effective HSP length: 98
effective length of query: 306
effective length of database: 5,391,079
effective search space: 1649670174
effective search space used: 1649670174
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0067b.6


