

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0066.6
(137 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
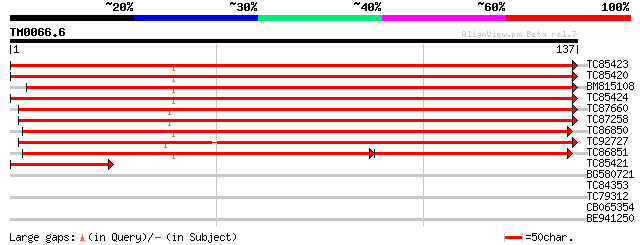
Score E
Sequences producing significant alignments: (bits) Value
TC85423 similar to SP|Q9FVI1|ADF2_PETHY Actin-depolymerizing fac... 253 2e-68
TC85420 similar to SP|Q9FVI1|ADF2_PETHY Actin-depolymerizing fac... 249 2e-67
BM815108 similar to SP|Q9FVI1|ADF2 Actin-depolymerizing factor 2... 246 3e-66
TC85424 similar to SP|Q9ZSK3|ADF4_ARATH Actin-depolymerizing fac... 232 3e-62
TC87660 similar to GP|18874466|gb|AAL79826.1 actin depolymerizin... 174 8e-45
TC87258 similar to GP|4566614|gb|AAD23407.1| actin depolymerizin... 169 3e-43
TC86850 similar to SP|Q9ZNT3|ADF5_ARATH Actin-depolymerizing fac... 166 3e-42
TC92727 weakly similar to GP|15149386|gb|AAK85273.1 cofilin {Pic... 112 4e-26
TC86851 similar to SP|Q9ZNT3|ADF5_ARATH Actin-depolymerizing fac... 103 2e-23
TC85421 homologue to SP|P51425|RL39_MAIZE 60S ribosomal protein ... 50 3e-07
BG580721 GP|4185511|gb| actin depolymerizing factor 4 {Arabidops... 30 0.30
TC84353 similar to GP|20466332|gb|AAM20483.1 putative protein {A... 27 3.3
TC79312 similar to GP|4567095|gb|AAD23584.1| fertilization-indep... 25 7.4
CB065354 weakly similar to PIR|D83400|D83 conserved hypothetical... 25 7.4
BE941250 25 9.7
>TC85423 similar to SP|Q9FVI1|ADF2_PETHY Actin-depolymerizing factor 2 (ADF
2). [Petunia] {Petunia hybrida}, partial (96%)
Length = 937
Score = 253 bits (646), Expect = 2e-68
Identities = 126/139 (90%), Positives = 132/139 (94%), Gaps = 2/139 (1%)
Frame = +3
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKL+FMELK KRT+RFIVYKIE+KQ VIVEKLGEP QGYEDFTA L
Sbjct: 147 MANAASGMAVHDDCKLKFMELKAKRTHRFIVYKIEEKQKQVIVEKLGEPAQGYEDFTACL 326
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYAVYDFEYLTE NVPKSRIFFI WSPDT+RVRSKMIYAS+K+RFKRELDGIQIE
Sbjct: 327 PADECRYAVYDFEYLTEENVPKSRIFFIGWSPDTARVRSKMIYASTKERFKRELDGIQIE 506
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDPTEMGLDVFKSRAN
Sbjct: 507 LQATDPTEMGLDVFKSRAN 563
>TC85420 similar to SP|Q9FVI1|ADF2_PETHY Actin-depolymerizing factor 2 (ADF
2). [Petunia] {Petunia hybrida}, partial (96%)
Length = 884
Score = 249 bits (636), Expect = 2e-67
Identities = 123/139 (88%), Positives = 133/139 (95%), Gaps = 2/139 (1%)
Frame = +1
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKLRFMELK KRT+RFI+YKIE+KQ VIVEKLGEP QGYE+F A L
Sbjct: 121 MANAASGMAVHDDCKLRFMELKAKRTHRFIIYKIEEKQKQVIVEKLGEPVQGYEEFAACL 300
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYAV+D+E++TEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE
Sbjct: 301 PADECRYAVFDYEFMTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 480
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDPTE+GLDVFKSRA+
Sbjct: 481 LQATDPTEIGLDVFKSRAS 537
>BM815108 similar to SP|Q9FVI1|ADF2 Actin-depolymerizing factor 2 (ADF 2).
[Petunia] {Petunia hybrida}, partial (93%)
Length = 583
Score = 246 bits (627), Expect = 3e-66
Identities = 122/135 (90%), Positives = 128/135 (94%), Gaps = 2/135 (1%)
Frame = +2
Query: 5 ASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANLPADE 62
ASGMAVHDDCKL+FMELK KRT+RFIVYKIE+KQ VIVEKLGEP QGYEDFTA LPADE
Sbjct: 65 ASGMAVHDDCKLKFMELKAKRTHRFIVYKIEEKQKQVIVEKLGEPAQGYEDFTACLPADE 244
Query: 63 CRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQAT 122
CRYAVYDFEYLTE NVPKSRIFFI WSPDT+RVRSKMIYAS+K+RFKRELDGIQIELQAT
Sbjct: 245 CRYAVYDFEYLTEENVPKSRIFFIGWSPDTARVRSKMIYASTKERFKRELDGIQIELQAT 424
Query: 123 DPTEMGLDVFKSRAN 137
DPTEMGLDVFKSRAN
Sbjct: 425 DPTEMGLDVFKSRAN 469
>TC85424 similar to SP|Q9ZSK3|ADF4_ARATH Actin-depolymerizing factor 4
(ADF-4) (AtADF4). [Mouse-ear cress] {Arabidopsis
thaliana}, complete
Length = 891
Score = 232 bits (592), Expect = 3e-62
Identities = 111/139 (79%), Positives = 127/139 (90%), Gaps = 2/139 (1%)
Frame = +2
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANL 58
MANAASGMAV+D+CKL+F+ELK KRTYR+I+YKIE+KQ V+V+K+G+P GY+DFTANL
Sbjct: 257 MANAASGMAVNDECKLKFLELKAKRTYRYIIYKIEEKQKQVVVDKVGDPANGYDDFTANL 436
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYAVYDF+++TE N KSRIFFIAW PD SRVRSKMIYASSKDRFKRELDGIQ+E
Sbjct: 437 PADECRYAVYDFDFVTEENCQKSRIFFIAWCPDISRVRSKMIYASSKDRFKRELDGIQVE 616
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDPTEM LDVFKSR N
Sbjct: 617 LQATDPTEMDLDVFKSRVN 673
>TC87660 similar to GP|18874466|gb|AAL79826.1 actin depolymerizing factor
{Vitis vinifera}, complete
Length = 798
Score = 174 bits (442), Expect = 8e-45
Identities = 77/137 (56%), Positives = 112/137 (81%), Gaps = 2/137 (1%)
Frame = +2
Query: 3 NAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANLPA 60
NA+SGM V D+ K FMELK K+ +R++++K+++K +V+VEK G P + Y+DF A+LP
Sbjct: 104 NASSGMGVDDNSKNTFMELKQKKVHRYVIFKVDEKKREVVVEKTGGPAESYDDFAASLPD 283
Query: 61 DECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQ 120
++CRYAV+DF+++T N KS+IFFIAWSP TSR+R+KM+YA++K+RF+RELDG+ E+Q
Sbjct: 284 NDCRYAVFDFDFVTAENCQKSKIFFIAWSPSTSRIRAKMLYATTKERFRRELDGVHYEIQ 463
Query: 121 ATDPTEMGLDVFKSRAN 137
ATDPTEM L+V + RA+
Sbjct: 464 ATDPTEMDLEVLRDRAH 514
>TC87258 similar to GP|4566614|gb|AAD23407.1| actin depolymerizing factor
{Populus x canescens}, complete
Length = 1013
Score = 169 bits (428), Expect = 3e-43
Identities = 77/137 (56%), Positives = 107/137 (77%), Gaps = 2/137 (1%)
Frame = +3
Query: 3 NAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANLPA 60
NA+SGM V + F EL+ K+ YR++++KI++K +V+VEK G P + Y+DFTA+LP
Sbjct: 216 NASSGMGVAEQSVSTFQELQRKKVYRYVIFKIDEKKKEVVVEKTGGPSESYDDFTASLPE 395
Query: 61 DECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQ 120
++CRYAV+DF+++T N KS+IFFIAWSP +R+R KM+YA+SKDRF+REL GI E+Q
Sbjct: 396 NDCRYAVFDFDFVTAENCQKSKIFFIAWSPSVARIRPKMLYATSKDRFRRELQGIHYEIQ 575
Query: 121 ATDPTEMGLDVFKSRAN 137
ATDPTEM L+V + RAN
Sbjct: 576 ATDPTEMELEVLQERAN 626
>TC86850 similar to SP|Q9ZNT3|ADF5_ARATH Actin-depolymerizing factor 5
(ADF-5) (AtADF5). [Mouse-ear cress] {Arabidopsis
thaliana}, complete
Length = 803
Score = 166 bits (420), Expect = 3e-42
Identities = 78/135 (57%), Positives = 104/135 (76%), Gaps = 2/135 (1%)
Frame = +2
Query: 4 AASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANLPAD 61
A +GM V D+CK FME+K K+ +R+IV+KI++K V V+K+G PG+ Y+D A+LP D
Sbjct: 104 ATTGMWVTDECKNSFMEMKWKKVHRYIVFKIDEKTRLVTVDKVGGPGENYDDLAASLPND 283
Query: 62 ECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQA 121
+CRYAV+DF+++T N KS+IFFIAWSP SR+R K++YA+SKD +R LDGI ELQA
Sbjct: 284 DCRYAVFDFDFVTVDNCRKSKIFFIAWSPTASRIREKILYATSKDGLRRALDGISYELQA 463
Query: 122 TDPTEMGLDVFKSRA 136
TDPTEMG DV + RA
Sbjct: 464 TDPTEMGFDVIQDRA 508
>TC92727 weakly similar to GP|15149386|gb|AAK85273.1 cofilin {Pichia
angusta}, partial (44%)
Length = 539
Score = 112 bits (281), Expect = 4e-26
Identities = 54/137 (39%), Positives = 91/137 (66%), Gaps = 2/137 (1%)
Frame = +3
Query: 3 NAASGMAVHDDCKLRFMELKTKRTYRFIVYKIED--KQVIVEKLGEPGQGYEDFTANLPA 60
+++SG+ + ++C F +LK K+ Y++I+YK++D K +++EK+ E Y+DF + L +
Sbjct: 30 SSSSGVDLSEECLEYFQDLKLKKKYKYILYKLDDSYKSIVLEKVVEEAT-YDDFISELTS 206
Query: 61 DECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQ 120
RYAVYDF+Y G +S+I F +W PD S+V+ KM+YASSKD ++ L G+ IE+Q
Sbjct: 207 SGPRYAVYDFDYEKPGEGQRSKIAFYSWIPDDSKVKDKMLYASSKDAIRKRLVGVAIEIQ 386
Query: 121 ATDPTEMGLDVFKSRAN 137
TD +E+ + +A+
Sbjct: 387 GTDLSEVSYEAVLEKAS 437
>TC86851 similar to SP|Q9ZNT3|ADF5_ARATH Actin-depolymerizing factor 5
(ADF-5) (AtADF5). [Mouse-ear cress] {Arabidopsis
thaliana}, complete
Length = 936
Score = 103 bits (258), Expect = 2e-23
Identities = 47/87 (54%), Positives = 67/87 (76%), Gaps = 2/87 (2%)
Frame = +2
Query: 4 AASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANLPAD 61
A +GM V D+CK FME+K K+ +R+IV+KI++K V V+K+G PG+ Y+D A+LP D
Sbjct: 143 ATTGMWVTDECKNSFMEMKWKKVHRYIVFKIDEKTRLVTVDKVGGPGENYDDLAASLPND 322
Query: 62 ECRYAVYDFEYLTEGNVPKSRIFFIAW 88
+CRYAV+DF+++T N KS+IFFIAW
Sbjct: 323 DCRYAVFDFDFVTVDNCRKSKIFFIAW 403
Score = 67.0 bits (162), Expect = 2e-12
Identities = 31/48 (64%), Positives = 37/48 (76%)
Frame = +2
Query: 89 SPDTSRVRSKMIYASSKDRFKRELDGIQIELQATDPTEMGLDVFKSRA 136
SP SR+R K++YA+SKD +R LDGI ELQATDPTEMG DV + RA
Sbjct: 764 SPTASRIREKILYATSKDGLRRALDGISYELQATDPTEMGFDVIQDRA 907
>TC85421 homologue to SP|P51425|RL39_MAIZE 60S ribosomal protein L39.
[Maize] {Zea mays}, complete
Length = 659
Score = 50.1 bits (118), Expect = 3e-07
Identities = 23/25 (92%), Positives = 23/25 (92%)
Frame = +2
Query: 1 MANAASGMAVHDDCKLRFMELKTKR 25
MANAASGMAVHDDCKLRFMEL KR
Sbjct: 584 MANAASGMAVHDDCKLRFMELXAKR 658
>BG580721 GP|4185511|gb| actin depolymerizing factor 4 {Arabidopsis
thaliana}, partial (9%)
Length = 563
Score = 30.0 bits (66), Expect = 0.30
Identities = 13/13 (100%), Positives = 13/13 (100%)
Frame = +2
Query: 1 MANAASGMAVHDD 13
MANAASGMAVHDD
Sbjct: 524 MANAASGMAVHDD 562
>TC84353 similar to GP|20466332|gb|AAM20483.1 putative protein
{Arabidopsis thaliana}, partial (9%)
Length = 602
Score = 26.6 bits (57), Expect = 3.3
Identities = 12/45 (26%), Positives = 26/45 (57%)
Frame = +1
Query: 7 GMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQVIVEKLGEPGQGY 51
G ++D C ++F + ++ ++ K+E+KQ I ++G +GY
Sbjct: 31 GRRINDGCMVKFSQKESIKSEDDNQMKLEEKQEIKTEMGLEVKGY 165
>TC79312 similar to GP|4567095|gb|AAD23584.1| fertilization-independent
endosperm protein {Arabidopsis thaliana}, partial (97%)
Length = 1480
Score = 25.4 bits (54), Expect = 7.4
Identities = 22/92 (23%), Positives = 38/92 (40%), Gaps = 10/92 (10%)
Frame = +1
Query: 29 FIVYKIEDKQVIV----EKLGEPGQGYEDFTANLPADECRY------AVYDFEYLTEGNV 78
FI+ K D ++I+ K PG+G D P EC + F+ GN
Sbjct: 961 FILSKSVDHEIILWEPKVKEQSPGEGAADILQKYPVPECDIWFIKFSCDFHFKACAIGN- 1137
Query: 79 PKSRIFFIAWSPDTSRVRSKMIYASSKDRFKR 110
+ +IF + +K+++A SK ++
Sbjct: 1138 REGKIFVWDLQSSPPILNAKLVHAQSKSPIRQ 1233
>CB065354 weakly similar to PIR|D83400|D83 conserved hypothetical protein
PA1972 [imported] - Pseudomonas aeruginosa (strain
PAO1), partial (29%)
Length = 626
Score = 25.4 bits (54), Expect = 7.4
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 3/51 (5%)
Frame = -3
Query: 55 TANLPAD---ECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYA 102
T +LP+ + R A+ F TEGN P SRI + + SR + ++ A
Sbjct: 312 TTSLPSSSRCQAR*AIGAFHSNTEGNTPSSRI*AKNLNDNKSRTSATLVSA 160
>BE941250
Length = 539
Score = 25.0 bits (53), Expect = 9.7
Identities = 9/22 (40%), Positives = 16/22 (71%)
Frame = -2
Query: 10 VHDDCKLRFMELKTKRTYRFIV 31
+H+ CK+R ++K K TY+ +V
Sbjct: 298 MHNICKVRTTDIKKKLTYKSLV 233
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,104,985
Number of Sequences: 36976
Number of extensions: 32335
Number of successful extensions: 192
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 182
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 183
length of query: 137
length of database: 9,014,727
effective HSP length: 86
effective length of query: 51
effective length of database: 5,834,791
effective search space: 297574341
effective search space used: 297574341
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0066.6


