

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
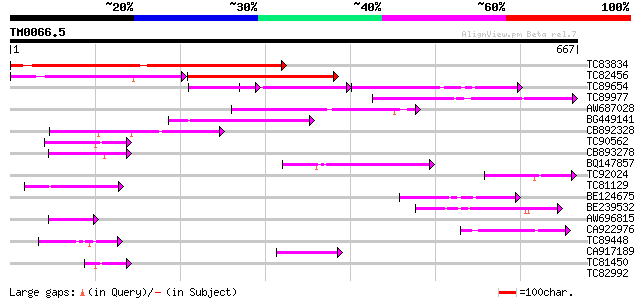
Query= TM0066.5
(667 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 509 e-144
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 181 5e-58
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 114 3e-40
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 141 9e-34
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 119 4e-27
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 95 8e-20
CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sin... 89 4e-18
TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 70 2e-12
CB893278 weakly similar to PIR|T05645|T056 hypothetical protein ... 68 1e-11
BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {A... 67 2e-11
TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10... 61 2e-09
TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 61 2e-09
BE124675 weakly similar to GP|13365573|db hypothetical protein~s... 59 6e-09
BE239532 59 8e-09
AW696815 homologue to GP|11994736|dbj far-red impaired response ... 52 6e-07
CA922976 weakly similar to GP|9502168|gb|A contains simlarity to... 52 1e-06
TC89448 similar to PIR|C84864|C84864 hypothetical protein At2g43... 50 4e-06
CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.3... 49 5e-06
TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 42 6e-04
TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D... 41 0.002
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 509 bits (1310), Expect = e-144
Identities = 243/325 (74%), Positives = 272/325 (82%)
Frame = +3
Query: 1 MEETSLCCEDLPHGECIEVHKDEDAALIELDSQNGFSEGRKEFDAPAVGMEFESYDDAYN 60
+++ S+C + LP EC +IELD QN SEGRKEF APA+ MEFESYDDAY+
Sbjct: 105 VDDISVCYDQLPDKEC---------QVIELDCQNDISEGRKEFPAPALAMEFESYDDAYS 257
Query: 61 YYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDVNHLRKETRTGCPAMM 120
YYICYAKEVGFCVRVKNSWFKRNS+EKYGAVLCCSSQGFKRTKDVN+LRKETRTGCPAM+
Sbjct: 258 YYICYAKEVGFCVRVKNSWFKRNSKEKYGAVLCCSSQGFKRTKDVNNLRKETRTGCPAMI 437
Query: 121 RIRIAESQRWRIIEVILEHNHILGAKIHKSAKKMGNGTKRKSLPSSDSEVQTVKLYRPLV 180
R+++ ESQRWRI EV LEHNH+LGAKIHKS KK SLPSSD+E +T+K+Y LV
Sbjct: 438 RMKLVESQRWRICEVTLEHNHVLGAKIHKSIKK-------NSLPSSDAEGKTIKVYHALV 596
Query: 181 IDAGGNGNLNPSARDDRTFSEMSNKLNVKRGDTQAIYNFLCRMQLTNPNFFYLMDFDDEG 240
ID GN NLN +ARDDR FS+ SNKLN+++GDTQAIYNFLCRMQLTNPNFFYLMDF+DEG
Sbjct: 597 IDTEGNDNLNSNARDDRAFSKYSNKLNLRKGDTQAIYNFLCRMQLTNPNFFYLMDFNDEG 776
Query: 241 HLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLLGCGLLAG 300
HLRNA W DA+SRAACGYF DVIYFDNTYL +KYEIPLV VGINHHGQSVLLGCGLLAG
Sbjct: 777 HLRNALWVDAKSRAACGYFSDVIYFDNTYLVNKYEIPLVALVGINHHGQSVLLGCGLLAG 956
Query: 301 ETTESYIWLFRTWVTCMSGCSPQTI 325
E ESY WLFRTW+ C+ CSPQTI
Sbjct: 957 EIIESYKWLFRTWIKCIPRCSPQTI 1031
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 181 bits (459), Expect(2) = 5e-58
Identities = 87/178 (48%), Positives = 120/178 (66%)
Frame = +2
Query: 210 RGDTQAIYNFLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTY 269
RGD AI++F RMQ N F+ MD DD+ +++N FWADAR RAA YFG+VI D TY
Sbjct: 710 RGDVDAIHSFFHRMQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDTTY 889
Query: 270 LSSKYEIPLVVFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDR 329
L+SKY++PLV FVG+NHHGQ++LLGC +L+ ++ WLF W+ CM G +P IIT+
Sbjct: 890 LTSKYDLPLVPFVGVNHHGQTILLGCAILSNLDAKTLTWLFTRWLECMHGHAPNGIITEE 1069
Query: 330 CEALQSAIIEVFPKSHHCFGLSLIMKKVPEKLGGLHNYDGIRKALIKAVYETLKISEF 387
+A+++AI FPK+ H + L IMKKVPE LG + I+ L VY+++ S+F
Sbjct: 1070DKAMKNAIEVAFPKARHRWCLWHIMKKVPEMLGKYSRNESIKTLLQDVVYDSMSKSDF 1243
Score = 62.4 bits (150), Expect(2) = 5e-58
Identities = 51/212 (24%), Positives = 89/212 (41%), Gaps = 4/212 (1%)
Frame = +1
Query: 1 MEETSLCCEDLPHGECIEVHKDEDAALIELDSQNGFSEGRKEFDAPAVGMEFESYDDAYN 60
MEE C+ + E + D+ +A I + E + P GM F S ++
Sbjct: 103 MEEIVASCDKVEKSEASKRSDDKFSAAIVVS----------EDEEPRTGMTFRSEEEVIR 252
Query: 61 YYICYAKEVGFCVRVKNSWFKRNSREKYGAVLC-CSSQGFKRTKDVNHLRKETRTGCPAM 119
YY+ YA VGF V K S + +KY + C C+ + +K+ + ++T C A
Sbjct: 253 YYMNYANRVGFGV-TKISSKNADDGKKYFTLACNCARRYVSTSKNPSKQYLTSKTQCRAR 429
Query: 120 MRIRIAESQRWRIIEVILEHNHILG---AKIHKSAKKMGNGTKRKSLPSSDSEVQTVKLY 176
+ +A I ++LEHNH LG + + + G +R + + +
Sbjct: 430 LNACVALDGSSTISRIVLEHNHELGQTKGRYFRLDESSGTHVERNLELNDQGGINVDRNL 609
Query: 177 RPLVIDAGGNGNLNPSARDDRTFSEMSNKLNV 208
+ + ++ G GNL D R F + +L +
Sbjct: 610 QSVGLEMNGCGNLTSGENDCRNFVQKVRRLRL 705
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 114 bits (285), Expect(2) = 3e-40
Identities = 68/203 (33%), Positives = 103/203 (50%), Gaps = 2/203 (0%)
Frame = +1
Query: 403 HEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLDKYELA 462
+EW +S+Y R W PV+L+ FFG + E + FF +V+ T L+ + +YE A
Sbjct: 697 NEWPQSMYNARQHWVPVYLRGSFFGEIPLNDGNECLNFFFDGHVNASTTLQLLVRQYEKA 876
Query: 463 LHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFG--TT 520
+ H+ E AD E+ NS+PVL+T +E Q + +YTR++FMKFQ EE+ T
Sbjct: 877 VSTWHERELKADFETSNSNPVLRTPSPMEKQAASLYTRKMFMKFQ---EELVGTMANPAT 1047
Query: 521 QLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCV 580
++ G I Y RV G + V ++ + + C C F + G +CRH L V
Sbjct: 1048KIEDSGTITTY----RVAKFGENQ--TSHTVAFNSSEMKASCSCQMFEYSGIVCRHILAV 1209
Query: 581 LNFNGVEEIPFKYILSRWKKDYK 603
V +P Y+L RW + K
Sbjct: 1210FRAKNVLTLPSHYVLKRWTMNAK 1278
Score = 80.9 bits (198), Expect = 1e-15
Identities = 36/85 (42%), Positives = 47/85 (54%)
Frame = +2
Query: 211 GDTQAIYNFLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYL 270
G + ++L RM+ N FFY + D + N FWAD R YFGD + FD TY
Sbjct: 116 GGGHQVLDYLKRMRAENHAFFYAVQSDVDNAGGNIFWADETCRTNYSYFGDTVIFDTTYK 295
Query: 271 SSKYEIPLVVFVGINHHGQSVLLGC 295
+++Y +P F G NHHGQ VL GC
Sbjct: 296 TNQYRVPFASFTGFNHHGQPVLFGC 370
Score = 70.1 bits (170), Expect(2) = 3e-40
Identities = 43/135 (31%), Positives = 72/135 (52%), Gaps = 3/135 (2%)
Frame = +3
Query: 271 SSKYEIPLVV--FVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITD 328
S+ Y +PL + + +N +VLL + E+ SYIWLF+TW+ +SG P +I TD
Sbjct: 303 STGYHLPLSLGLIIMVNLFYLAVLL----ILNESEPSYIWLFKTWLRAVSGRPPVSITTD 470
Query: 329 RCEALQSAIIEVFPKSHHCFGLSLIMKKVPEKLGGLHNYD-GIRKALIKAVYETLKISEF 387
+Q A+ +V P + H F I ++ KL L+ + K V+E++ I EF
Sbjct: 471 LDPVIQVAVAQVLPPTRHRFCKWSIFRENRSKLAHLYQSNPTFDNEFKKCVHESVTIDEF 650
Query: 388 EAAWGIMIQRFGVSD 402
E+ W +++R+ + D
Sbjct: 651 ESCWRSLLERYYIMD 695
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 141 bits (355), Expect = 9e-34
Identities = 84/240 (35%), Positives = 127/240 (52%)
Frame = +2
Query: 428 GMSAARPGESIGPFFGRYVHKQTPLKEFLDKYELALHKKHKEESLADTESRNSSPVLKTR 487
G S A+ ES+ FF +Y+HK+ LKEF+ +Y L L ++ EE +AD ++ + P LK+
Sbjct: 5 GTSTAQRSESMNSFFDKYIHKKITLKEFVKQYRLILLNRYDEEEIADFDTLHKQPALKSP 184
Query: 488 CSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQLHVDGPIVIYLVKERVLIDGNRREIK 547
E Q+S +YT IF KFQ EV + C ++ DG V Y+V++ + +
Sbjct: 185 SPWEKQMSTIYTHAIFKKFQVEVLGVAGCQSRIEVG-DGTAVRYIVQD-------YEKDE 340
Query: 548 DFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYKRLHV 607
+F V + + E+ C C F + G+LCRHAL VL G +P YI+ RW KD K +
Sbjct: 341 EFLVTWKELSSEVSCFCRLFEYKGFLCRHALSVLQRCGCSSVPSHYIMKRWTKDAK-IRE 517
Query: 608 PDHSSVSADDTDPIQWSNQLFSSALQVVEEGTISLDHYNVALQACEESLSKVHDVDQRQD 667
P T P Q N L ++ + EEG+ S + YN+AL+A ++L V+ D
Sbjct: 518 PAADRTQRIQTRP-QRYNDLCKRSIALSEEGSSSEESYNIALRALIDALKNCVLVNNSND 694
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 119 bits (298), Expect = 4e-27
Identities = 70/231 (30%), Positives = 108/231 (46%), Gaps = 8/231 (3%)
Frame = +1
Query: 261 DVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGC 320
DV+ FD TY +KY PL +F G NHH Q+++ G LL ET ESY W+ ++ CM
Sbjct: 1 DVLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENK 180
Query: 321 SPQTIITDRCEALQSAIIEVFPKSHHCFGLSLIMKKVPEKLGGLHNYDGIRKALIKAVYE 380
P+ ++TD +++ AI +VFP + H + K E + +G RKA+ Y
Sbjct: 181 FPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENIKKTPFLEGFRKAM----YS 348
Query: 381 TLKISEFEAAWGIMIQRFGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGP 440
+FE W +IQ+ + + W+ Y ++ WA +L+D FFG + E+I
Sbjct: 349 NFTPEQFEDFWSELIQKNELEGNAWVIKTYANKSLWATAYLRDKFFGRIRTTSQCEAINA 528
Query: 441 FFGRYVHKQTP--------LKEFLDKYELALHKKHKEESLADTESRNSSPV 483
Y Q L F + Y E +AD +S+ + PV
Sbjct: 529 IVKTYSRSQRQNF*IYA*FLNRF*EGY-------RNNELVADFKSKFTEPV 660
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 95.1 bits (235), Expect = 8e-20
Identities = 53/172 (30%), Positives = 89/172 (50%)
Frame = +3
Query: 187 GNLNPSARDDRTFSEMSNKLNVKRGDTQAIYNFLCRMQLTNPNFFYLMDFDDEGHLRNAF 246
G L + +D R + KL+ + +T + ++ +PNF + D L N
Sbjct: 159 GYLPFTEKDVRNLLQSFRKLDPEE-ETLDLLRMCRNIKDKDPNFKFEYTLDANNRLENIA 335
Query: 247 WADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLLGCGLLAGETTESY 306
W+ A S FGD + FD T+ + +++PL ++VGIN++G GC LL ET S+
Sbjct: 336 WSYASSIQLYDIFGDAVVFDTTHRLTAFDMPLGLWVGINNYGMPCFFGCVLLRDETVRSF 515
Query: 307 IWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPKSHHCFGLSLIMKKVP 358
W + ++ M+G +PQTI+TD+ L+ A+ P + H F + +I+ K P
Sbjct: 516 SWAIKAFLGFMNGKAPQTILTDQNICLKEALSAEMPMTKHAFCIWMIVAKFP 671
>CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sinorhizobium
meliloti}, partial (1%)
Length = 828
Score = 89.4 bits (220), Expect = 4e-18
Identities = 62/215 (28%), Positives = 100/215 (45%), Gaps = 10/215 (4%)
Frame = +1
Query: 48 VGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAV-LCCSSQGFKRTK--- 103
+G S D+AY+ Y +A + GF VR + N ++K CS QGFK +
Sbjct: 160 LGQVVNSKDEAYDLYQEHAFKTGFSVRKGKELYYDNEKKKTRLKDFYCSKQGFKNNEPDG 339
Query: 104 DVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNH----ILGAKIHKSAKKMGN--G 157
+V + R ++RT C AM+R + + W++ ++IL+HNH + + +S + M N
Sbjct: 340 EVAYKRADSRTNCLAMVRFNVTKEGVWKVTKLILDHNHEFVPLQQRYLLRSMRNMSNLKE 519
Query: 158 TKRKSLPSSDSEVQTVKLYRPLVIDAGGNGNLNPSARDDRTFSEMSNKLNVKRGDTQAIY 217
+ KSL + V V Y L + GG +D + ++ GD Q++
Sbjct: 520 DRIKSLVNDAIRVTNVGGY--LGKEVGGVDKRGVMLKDMHNYVSTGKLKFIEAGDAQSLL 693
Query: 218 NFLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARS 252
N L Q + F+Y + D E L N FW D +S
Sbjct: 694 NHLQSKQAQDSMFYYSVQLDHESRLNNVFWRDGKS 798
>TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (66%)
Length = 1169
Score = 70.5 bits (171), Expect = 2e-12
Identities = 41/107 (38%), Positives = 53/107 (49%), Gaps = 4/107 (3%)
Frame = +2
Query: 41 KEFDAPAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGF- 99
+ D P +G EF S +A+ +Y YA VGF +RV R G L C+ +GF
Sbjct: 308 RTIDEPYIGQEFVSEAEAHAFYNSYATRVGFVIRVSKLSRSRRDGSVIGRALVCNKEGFR 487
Query: 100 ---KRTKDVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHIL 143
KR K V R ETR GC AM+ +R S W I + + EH H L
Sbjct: 488 MPDKREKIVRQ-RAETRVGCRAMIMVRKLNSGLWSITKFVKEHTHPL 625
>CB893278 weakly similar to PIR|T05645|T056 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (5%)
Length = 821
Score = 67.8 bits (164), Expect = 1e-11
Identities = 33/104 (31%), Positives = 56/104 (53%), Gaps = 6/104 (5%)
Frame = +3
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDV 105
P +GM+F S ++A +Y Y + +GF VR+ ++ R + E G CS +GF+ K V
Sbjct: 315 PFIGMQFNSREEARGFYDGYGRRIGFTVRIHHNRRSRVNNELIGQDFVCSKEGFRAKKYV 494
Query: 106 NHLRK------ETRTGCPAMMRIRIAESQRWRIIEVILEHNHIL 143
+ + TR GC AM+R+ + + +W + + + EH H L
Sbjct: 495 HRKDRVLPPPPATREGCQAMIRLALRDEGKWVVTKFVKEHTHKL 626
>BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (26%)
Length = 560
Score = 67.0 bits (162), Expect = 2e-11
Identities = 50/183 (27%), Positives = 84/183 (45%), Gaps = 4/183 (2%)
Frame = +1
Query: 321 SPQTIITDRCEALQSAIIEVFPKSHHCFGLSLIMKKVPE----KLGGLHNYDGIRKALIK 376
+PQT++TD L+ AI P+S H F + I+ K + LG YD KA
Sbjct: 1 APQTLLTDHNTWLKEAIAVEMPESKHAFCIWHILSKFSDWXYLLLGS--QYDEW-KADFH 171
Query: 377 AVYETLKISEFEAAWGIMIQRFGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGE 436
+Y +FE +W M+ ++G+ ++ + SLY R WA FL+ FF G+++ E
Sbjct: 172 RLYNLEMXEDFEKSWRQMVDKYGLHANKHIISLYSLRTFWALPFLRRYFFAGLTSXCQTE 351
Query: 437 SIGPFFGRYVHKQTPLKEFLDKYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSR 496
SI F R++ Q+ FL + + + + + + LKT +E +
Sbjct: 352 SINVFIQRFLSAQSQPXRFLQQVADIVDFNDRAGAKQXMQRKMQKXCLKTGSPIESHAAT 531
Query: 497 MYT 499
+ T
Sbjct: 532 ILT 540
>TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (11%)
Length = 938
Score = 60.8 bits (146), Expect = 2e-09
Identities = 38/114 (33%), Positives = 62/114 (54%), Gaps = 6/114 (5%)
Frame = +3
Query: 559 ELRCICSC--FNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYK-RLHVPDHSSVSA 615
E++ CSC F F G LCRH L V V +P YIL RW K+ K + + +HSS +
Sbjct: 78 EMKATCSCQMFEFSGLLCRHILAVFRVTNVLTLPSHYILKRWTKNAKSNVSLQEHSSHAY 257
Query: 616 D---DTDPIQWSNQLFSSALQVVEEGTISLDHYNVALQACEESLSKVHDVDQRQ 666
++ +++ N L A + V++G S + Y+VA A +E+ +V V +++
Sbjct: 258 TYYLESHTVRY-NTLRHEAFKFVDKGASSPETYDVAKDALQEAAKRVAQVMRKE 416
>TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (13%)
Length = 721
Score = 60.8 bits (146), Expect = 2e-09
Identities = 36/119 (30%), Positives = 57/119 (47%), Gaps = 2/119 (1%)
Frame = +2
Query: 18 EVHKDEDAALIELDSQNGFSEGRKEFDAPAVGMEFESYDDAYNYYICYAKEVGFCVRVKN 77
E H A + F EG +F+ P G+EFE+++ AY++Y YAK +GF +KN
Sbjct: 332 EEHHTSGALVASPKRGIAFFEGDTDFEPPN-GIEFETHEAAYSFYQEYAKSMGFTTSIKN 508
Query: 78 SWFKRNSREKYGAVLCCSSQGFKRTKDVNHLRKET--RTGCPAMMRIRIAESQRWRIIE 134
S + ++E A CS G D ++ + +T C A M ++ +W I E
Sbjct: 509 SRRSKKTKEFIDAKFACSRYGVTPESDGRSSQRSSVKKTDCKACMHVKRNPDGKWNIFE 685
>BE124675 weakly similar to GP|13365573|db hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (4%)
Length = 448
Score = 58.9 bits (141), Expect = 6e-09
Identities = 42/143 (29%), Positives = 65/143 (45%)
Frame = +2
Query: 459 YELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFG 518
+E + ++ +E A E + P L + S YT F FQ E+ +
Sbjct: 2 FERIVEEQRYKEIEASDEMKGCLPKLMGNVVVLKHASVAYTPRAFEVFQQRYEKSLNVI- 178
Query: 519 TTQLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHAL 578
Q DG YL + +V G+ R+ + V +S + + C C F G+LC HAL
Sbjct: 179 VNQHKRDG----YLFEYKVNTYGHARQ---YTVTFSSSDNTVVCSCMKFEHVGFLCSHAL 337
Query: 579 CVLNFNGVEEIPFKYILSRWKKD 601
VL+ ++ +P +YIL RW KD
Sbjct: 338 KVLDNRNIKVVPSRYILKRWTKD 406
>BE239532
Length = 645
Score = 58.5 bits (140), Expect = 8e-09
Identities = 51/184 (27%), Positives = 83/184 (44%), Gaps = 11/184 (5%)
Frame = +3
Query: 478 RNSSPVLKTRCSLELQ-LSRMYTREIFMKFQFEVEEMFSCFGTTQLHVDGPIVIYLVKER 536
R S+P L S L+ S +YT + F F E E G+T + + L
Sbjct: 3 RQSAPGLVINISETLRHASTVYTHKGFRIFLNEYLE--GTGGSTSIEISQSN--NLSNHE 170
Query: 537 VLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIPFKYILS 596
V++ N + K + V + + ++ C C F+ G LC HAL + N G+ IP +Y L
Sbjct: 171 VML--NHKPNKKYAVTFDSSNMKINCSCRKFDSMGILCSHALRIYNIKGILRIPDQYFLK 344
Query: 597 RWKKDYKRL---HVP-------DHSSVSADDTDPIQWSNQLFSSALQVVEEGTISLDHYN 646
RW K+ + + H+P +SV DD I + N + +S +V E S + N
Sbjct: 345 RWSKNARSVIYEHMPRGMGEDSTSNSVINDDDAGILYRNAIMTSFYYLVLESQDSKEAQN 524
Query: 647 VALQ 650
+ ++
Sbjct: 525 IYVE 536
>AW696815 homologue to GP|11994736|dbj far-red impaired response protein;
Mutator-like transposase-like protein; phytochrome A
signaling, partial (7%)
Length = 394
Score = 52.4 bits (124), Expect = 6e-07
Identities = 26/59 (44%), Positives = 35/59 (59%)
Frame = +2
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKD 104
P GMEFES+ +AY++Y YA+ +GF ++NS + SRE A CS G KR D
Sbjct: 215 PLSGMEFESHGEAYSFYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYD 391
>CA922976 weakly similar to GP|9502168|gb|A contains simlarity to Arabidopsis
thaliana far-red impaired response protein
(GB:AAD51282.1), partial (11%)
Length = 827
Score = 51.6 bits (122), Expect = 1e-06
Identities = 32/129 (24%), Positives = 61/129 (46%)
Frame = -3
Query: 531 YLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIP 590
Y+++ +DG R V++ ++ C C F G LCRHAL +L ++P
Sbjct: 777 YIIRHSKKLDGERH------VLWLPEKEQILCSCKEFESSGILCRHALRILVTKNYFQLP 616
Query: 591 FKYILSRWKKDYKRLHVPDHSSVSADDTDPIQWSNQLFSSALQVVEEGTISLDHYNVALQ 650
KY LSRW ++ + +H++ S +W + S A + E +I+ + + A +
Sbjct: 615 EKYYLSRWHRERSLIVEDEHNNQSFIAE---KWFQEYQSQAENLFSESSITKERSDYARK 445
Query: 651 ACEESLSKV 659
+ L+++
Sbjct: 444 ELMKELTRI 418
>TC89448 similar to PIR|C84864|C84864 hypothetical protein At2g43280
[imported] - Arabidopsis thaliana, partial (80%)
Length = 1177
Score = 49.7 bits (117), Expect = 4e-06
Identities = 38/109 (34%), Positives = 52/109 (46%), Gaps = 11/109 (10%)
Frame = +1
Query: 35 GFSEGRKEFDAPAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVL-- 92
G S+ P GMEF S D A +Y YA+ VGF +RV + +R+ R+ G +L
Sbjct: 268 GISDHDNAVKEPYEGMEFVSEDAAKIFYDEYARHVGFVMRVMSC--RRSXRD--GRILAR 435
Query: 93 ---------CCSSQGFKRTKDVNHLRKETRTGCPAMMRIRIAESQRWRI 132
C S QG + V R TR GC AM+ I+ +S +W I
Sbjct: 436 RFGCNKEGHCVSIQG--KLGPVRKPRASTREGCKAMIHIKYDQSGKWMI 576
>CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (12%)
Length = 312
Score = 49.3 bits (116), Expect = 5e-06
Identities = 30/79 (37%), Positives = 40/79 (49%), Gaps = 1/79 (1%)
Frame = +1
Query: 314 VTCMSGCSPQTIITDRCEALQSAIIEVFPKSHHCFGLSLIMKKVPEKLGGLH-NYDGIRK 372
+T MSG P +I TD +QSAI++VFP + H F I K+ EKL + +
Sbjct: 1 LTAMSGRPPLSITTDHDSVIQSAIMQVFPDTRHRFCKWHIFKQCQEKLSHIFLQFPNFEA 180
Query: 373 ALIKAVYETLKISEFEAAW 391
K V T I EFE+ W
Sbjct: 181 EFHKCVNLTDSIDEFESCW 237
>TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (59%)
Length = 832
Score = 42.4 bits (98), Expect = 6e-04
Identities = 24/59 (40%), Positives = 32/59 (53%), Gaps = 4/59 (6%)
Frame = +2
Query: 89 GAVLCCSSQGF----KRTKDVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHIL 143
G L C+ +G+ KR K V R ETR GC AM+ +R S +W I + + EH H L
Sbjct: 167 GRALVCNKEGYRMPDKREKIVRQ-RAETRVGCRAMIMMRKINSGKWVITKFVKEHTHPL 340
>TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (7%)
Length = 354
Score = 40.8 bits (94), Expect = 0.002
Identities = 24/58 (41%), Positives = 36/58 (61%), Gaps = 4/58 (6%)
Frame = +1
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVL----CCSSQGF 99
P+ GMEFES + A +Y YA+ VGF RV +S +R+ R+ GA++ C+ +GF
Sbjct: 193 PSEGMEFESEEAAKAFYNSYARRVGFSTRVSSS--RRSRRD--GAIIQRQXVCAKEGF 354
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,326,528
Number of Sequences: 36976
Number of extensions: 359037
Number of successful extensions: 1775
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 1750
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1764
length of query: 667
length of database: 9,014,727
effective HSP length: 102
effective length of query: 565
effective length of database: 5,243,175
effective search space: 2962393875
effective search space used: 2962393875
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0066.5


