

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.20
(196 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
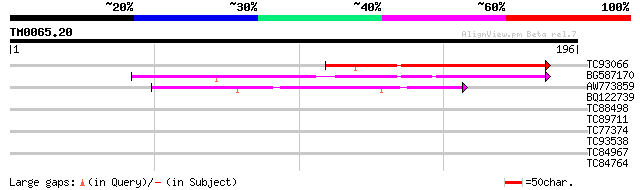
Score E
Sequences producing significant alignments: (bits) Value
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 74 5e-14
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 67 3e-12
AW773859 44 5e-05
BQ122739 34 0.042
TC88498 similar to GP|21304712|gb|AAM45475.1 ethylene-responsive... 33 0.071
TC89711 weakly similar to PIR|T10725|T10725 protein kinase Xa21 ... 29 1.0
TC77374 weakly similar to GP|14517446|gb|AAK62613.1 AT5g14030/MU... 29 1.3
TC93538 similar to SP|Q12644|NUIM_NEUCR NADH-ubiquinone oxidored... 28 2.3
TC84967 similar to PIR|G86453|G86453 YUP8H12R.39. homolog F9L11.... 28 3.0
TC84764 similar to PIR|S71584|S71584 GTP-binding protein BG1 - A... 27 5.1
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 73.6 bits (179), Expect = 5e-14
Identities = 38/79 (48%), Positives = 52/79 (65%), Gaps = 1/79 (1%)
Frame = +1
Query: 110 LGVCEHCIL-GKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDST 168
L C+H + G ++KVSFS A ++K L +H+D+WGP+ V S GG RY +T IDD
Sbjct: 13 LEFCKHLLFFGNRKKVSFSTATHRTK-GILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFP 189
Query: 169 RKV*VYFLKNKSDVFSVFK 187
RKV VYFL+ K++ F FK
Sbjct: 190 RKVWVYFLRYKNETFPTFK 246
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 67.4 bits (163), Expect = 3e-12
Identities = 42/152 (27%), Positives = 73/152 (47%), Gaps = 7/152 (4%)
Frame = -3
Query: 43 KVTKGNLVVARGKKRGSLYMVAEEDMIA-------VTEAVNSSSIWHQRLGHMSEKGMKI 95
+V + + ++ G +G LYM+ + D ++ + ++N ++WH RLGH + + +
Sbjct: 716 EVIESSQLIGEGVTKGDLYMLEKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRALNL 537
Query: 96 MASKGKMSNLKHVDLGVCEHCILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLG 155
M N CE CILGK K F + + L++TD+W AP S
Sbjct: 536 MLPGVVFENKN------CEACILGKHCKNVFPRTSTVYE-NCFDLIYTDLW-TAPSLSRD 381
Query: 156 GSRYYVTFIDDSTRKV*VYFLKNKSDVFSVFK 187
+Y+VTFID+ ++ + + +K V FK
Sbjct: 380 NHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFK 285
>AW773859
Length = 538
Score = 43.5 bits (101), Expect = 5e-05
Identities = 30/122 (24%), Positives = 53/122 (42%), Gaps = 13/122 (10%)
Frame = -3
Query: 50 VVARGKKRGSLYMVAEEDMIAVTEAVNS------------SSIWHQRLGHMSEKGMKIMA 97
++ G+ LY + +D + T A + ++WH RLGH+S + K+++
Sbjct: 356 MIGSGELFEGLYFLTTQDTPSATIASSQVQPQPQPTFLPQEALWHFRLGHLSNR--KLLS 183
Query: 98 SKGKMSNLKHVDLGVCEHCILGKQRKVSFS-KAGRKSKLEKLKLVHTDVWGPAPVKSLGG 156
+ VC+ C + +K+ F R SK +L H D+WGP +S+
Sbjct: 182 LHSNFPFITIDQNSVCDICHYSRHKKLPFQLSTNRASKC--YELFHFDIWGPFSTQSIHN 9
Query: 157 SR 158
R
Sbjct: 8 QR 3
>BQ122739
Length = 575
Score = 33.9 bits (76), Expect = 0.042
Identities = 21/81 (25%), Positives = 40/81 (48%), Gaps = 6/81 (7%)
Frame = +1
Query: 28 LDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMIAVTEAVN------SSSIW 81
LDD+GY G ++T ++V+ +GK L ++ + +A+ SS
Sbjct: 223 LDDQGYIFNIEDGDLRITNDSMVLMKGKLENGLSLL*GRTSMDTADAIYIRCNLVSSRSS 402
Query: 82 HQRLGHMSEKGMKIMASKGKM 102
R+GH+S+ G+K + + G +
Sbjct: 403 AYRMGHVSKGGIKKLNTIGAL 465
>TC88498 similar to GP|21304712|gb|AAM45475.1 ethylene-responsive element
binding protein 1 {Glycine max}, partial (45%)
Length = 902
Score = 33.1 bits (74), Expect = 0.071
Identities = 29/93 (31%), Positives = 38/93 (40%), Gaps = 3/93 (3%)
Frame = +3
Query: 65 EEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMAS-KGKMSNLKHVDLG--VCEHCILGKQ 121
E D I VN+ IW + G K A + N V LG V E
Sbjct: 204 ESDTIVEAREVNTPPIWKRYKGVRRRPWGKFAAEIRDPKKNGARVWLGTYVTEEEAALAY 383
Query: 122 RKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSL 154
K +F GRK+KL L+ +DV+ P P K +
Sbjct: 384 DKAAFKMRGRKAKLNFPHLIGSDVFTPEPEKEV 482
>TC89711 weakly similar to PIR|T10725|T10725 protein kinase Xa21 (EC
2.7.1.-) A1 receptor type - long-staminate rice,
partial (7%)
Length = 1391
Score = 29.3 bits (64), Expect = 1.0
Identities = 12/25 (48%), Positives = 16/25 (64%)
Frame = -2
Query: 124 VSFSKAGRKSKLEKLKLVHTDVWGP 148
+S K+ E L+L+HTDVWGP
Sbjct: 910 ISPFKSSTSHAQEVLELIHTDVWGP 836
>TC77374 weakly similar to GP|14517446|gb|AAK62613.1 AT5g14030/MUA22_2
{Arabidopsis thaliana}, partial (88%)
Length = 950
Score = 28.9 bits (63), Expect = 1.3
Identities = 27/83 (32%), Positives = 35/83 (41%), Gaps = 1/83 (1%)
Frame = +1
Query: 112 VCEHCILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYY-VTFIDDSTRK 170
+C H V F A +K+ L +LK V + + G S Y V+ DDS
Sbjct: 130 LCSHA----SSDVPFIVAHKKASLNRLKTGAERVSVTIDIYNQGTSTAYDVSLSDDSWT- 294
Query: 171 V*VYFLKNKSDVFSVFKGTTKKT 193
SD+FSV GTT KT
Sbjct: 295 ---------SDLFSVISGTTSKT 336
>TC93538 similar to SP|Q12644|NUIM_NEUCR NADH-ubiquinone oxidoreductase 23
kDa subunit mitochondrial precursor (EC 1.6.5.3) (EC
1.6.99.3), partial (68%)
Length = 634
Score = 28.1 bits (61), Expect = 2.3
Identities = 15/30 (50%), Positives = 19/30 (63%)
Frame = +3
Query: 136 EKLKLVHTDVWGPAPVKSLGGSRYYVTFID 165
EKL L + D W P K+L ++YYV FID
Sbjct: 453 EKL-LANGDKWEPEIAKNLEANQYYV*FID 539
>TC84967 similar to PIR|G86453|G86453 YUP8H12R.39. homolog F9L11.9
[imported] - Arabidopsis thaliana, partial (20%)
Length = 515
Score = 27.7 bits (60), Expect = 3.0
Identities = 13/28 (46%), Positives = 19/28 (67%)
Frame = -1
Query: 113 CEHCILGKQRKVSFSKAGRKSKLEKLKL 140
CEH +L + K+SF++ R K+E LKL
Sbjct: 476 CEHAMLLVRNKLSFNRFIRVIKIEALKL 393
>TC84764 similar to PIR|S71584|S71584 GTP-binding protein BG1 - Arabidopsis
thaliana, partial (79%)
Length = 705
Score = 26.9 bits (58), Expect = 5.1
Identities = 17/64 (26%), Positives = 35/64 (54%)
Frame = +1
Query: 124 VSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYFLKNKSDVF 183
+ KAG+ + LEK+K V+T+V G P + + + I+ + RK+ + L + +
Sbjct: 283 LGIDKAGKTTLLEKIKSVYTNVEGLPPDRIVPTVGLNIGRIEVANRKLVFWDLGGQLGLR 462
Query: 184 SVFK 187
S+++
Sbjct: 463 SIWE 474
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,036,814
Number of Sequences: 36976
Number of extensions: 76643
Number of successful extensions: 313
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 312
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 312
length of query: 196
length of database: 9,014,727
effective HSP length: 91
effective length of query: 105
effective length of database: 5,649,911
effective search space: 593240655
effective search space used: 593240655
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0065.20


