

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
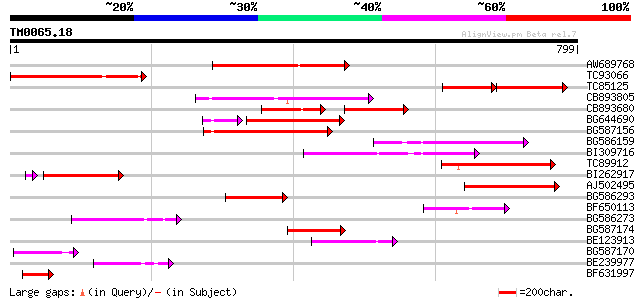
Query= TM0065.18
(799 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 186 2e-47
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 165 7e-41
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 93 3e-40
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 158 7e-39
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 90 2e-38
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 137 2e-38
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 155 6e-38
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 146 3e-35
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 146 3e-35
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 141 9e-34
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 135 2e-32
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 114 2e-25
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 88 1e-17
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 87 3e-17
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 84 2e-16
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 81 1e-15
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 78 1e-14
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 63 4e-10
BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryz... 62 1e-09
BF631997 weakly similar to GP|18542925|gb Putative pol polyprote... 44 3e-04
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 186 bits (473), Expect = 2e-47
Identities = 96/193 (49%), Positives = 125/193 (64%), Gaps = 1/193 (0%)
Frame = +1
Query: 287 KWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGS-RRYKARLVVKGFQ 345
+W AMK E +L N TW L LP KKA+ +WVYR+KE DGS ++KARLV KGF
Sbjct: 94 RWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFS 273
Query: 346 QKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGF 405
Q G D+TE FSPV+K TIR+IL+I ++Q+D+ AFL+G L+EE+YM+QP+GF
Sbjct: 274 QTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGF 453
Query: 406 EVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIIL 465
E K+LVCKL+KSLYGLKQAPR WY+ GF + D + + I L
Sbjct: 454 EA-ANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYL 630
Query: 466 ALYVDDMLIAGSN 478
+YVDD+LI GS+
Sbjct: 631 XIYVDDILITGSS 669
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 165 bits (417), Expect = 7e-41
Identities = 89/195 (45%), Positives = 125/195 (63%), Gaps = 4/195 (2%)
Frame = +1
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
W+ VE QT +K L +DN E+ S +F +FC+ +GI KTI P+QNGVAERM RT
Sbjct: 250 WRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRT 429
Query: 62 LNERARCMRIQSGL--PKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLK 119
L ERARCM +GL + WV+A +TA +L+NR P LD+++PE++WSG V S+L+
Sbjct: 430 LLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLR 609
Query: 120 VFGCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFW--DEQNKKIIRSRNVTFNES 177
+FGC +Y L++ KL P+A +C F+ Y + GYR W D +++K+I SR+VTFNE
Sbjct: 610 IFGCPAYALVND---GKLAPRAGECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNED 780
Query: 178 VLYKDRSSAESMSSS 192
L S +S SS
Sbjct: 781 ALLS--SGKQSFVSS 819
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 92.8 bits (229), Expect(2) = 3e-40
Identities = 45/76 (59%), Positives = 58/76 (76%)
Frame = +1
Query: 611 VKWILRYLKGSSRMCLCFRRNNLTLQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSK 670
VK I+RY+KG+S + +CF + LT++ + D+D GD D KSTTGY+FTL G AVSW SK
Sbjct: 1 VKRIMRYIKGTSGVAVCFGGSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSK 180
Query: 671 LQNRVALSTTESEYVA 686
LQ VALSTTE+EY+A
Sbjct: 181 LQTVVALSTTEAEYMA 228
Score = 91.7 bits (226), Expect(2) = 3e-40
Identities = 39/100 (39%), Positives = 67/100 (67%)
Frame = +3
Query: 686 AISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRE 745
++ +A KE IW++ ++ELG +Q+ ++ DSQS + +A+NP FHSR KHI ++YHF+RE
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVRE 407
Query: 746 LISDEELSLLKILGSENPTDMLTKTVTADNLRLCIASAGL 785
++ + + + KI ++N D +TK++ D C +S GL
Sbjct: 408 VVEEGSVDMQKIHTNDNLADAMTKSINTDKFIWCRSSYGL 527
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 158 bits (400), Expect = 7e-39
Identities = 84/255 (32%), Positives = 144/255 (55%), Gaps = 5/255 (1%)
Frame = +3
Query: 263 NYLLLTDAGEPEYFGEAMQGNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWV 322
N +LT +P F EA++ S KW +M +EM + ++N TW LT L G K + +W+
Sbjct: 12 NLGMLTMTSDPTTFEEAVK---SEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWI 182
Query: 323 YRLKEESDGS-RRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQ 381
++ K +G +YKARLV KG+ Q+ G+D+TE+F+PV + TIR+++++ A +++
Sbjct: 183 FKTKLNENGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAA----QIKR 350
Query: 382 LDVKIAFL---HGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEF 438
V I+ + H +E + V+ + + ++ ++LYGLKQAPR WY + +
Sbjct: 351 DGVCIS*M*KAHSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAY 530
Query: 439 MSNSGFNRCDMDHCCFVK-KFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKD 497
+ GF +C +H FVK +I++LYVDD++ G++ K+ M + F M D
Sbjct: 531 FTKEGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSD 710
Query: 498 LGPAKQILGMRISRN 512
LG LG+ +++N
Sbjct: 711 LGKMHYFLGVEVTQN 755
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 90.1 bits (222), Expect(2) = 2e-38
Identities = 51/89 (57%), Positives = 59/89 (65%)
Frame = -2
Query: 356 FSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVC 415
F P+VK+ TI +LSIVA ENL+LE LDVK AFL GDL E+IYM QPEGF K +V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGK-MVG 378
Query: 416 KLHKSLYGLKQAPRQWYKKFNEFMSNSGF 444
KL KS+YGLKQ PRQ + GF
Sbjct: 377 KLKKSMYGLKQGPRQCI*SLKALCTRKGF 291
Score = 88.6 bits (218), Expect(2) = 2e-38
Identities = 43/90 (47%), Positives = 65/90 (71%)
Frame = -3
Query: 472 MLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLD 531
+L+ GSN+ EI LK + S+ +MKDLGPAK+I+GM+I ++ +GVL LSQ +Y+ ++L
Sbjct: 271 LLVVGSNIDEIKNLKTRFSKEIDMKDLGPAKKIIGMQIMIDKQKGVL*LSQVEYITRVLQ 92
Query: 532 RFNVGDANTRSTPLGNHLKFSKKQSPQTDE 561
FN+G+A ST L +H S +QSPQT++
Sbjct: 91 IFNMGNAILVSTTLASHFCLSHEQSPQTEK 2
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 137 bits (345), Expect(2) = 2e-38
Identities = 64/139 (46%), Positives = 98/139 (70%)
Frame = -2
Query: 334 RYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDL 393
R K++LVV+G+ QK+GID+ E FSPV +M IR++++ A L Q+DVK AF++GDL
Sbjct: 418 RNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFINGDL 239
Query: 394 EEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCC 453
+EE+++ QP GFE N V +L+K+LYGLKQAPR WY++ ++F+ +GF R +D+
Sbjct: 238 KEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDNTL 59
Query: 454 FVKKFADSYIILALYVDDM 472
F+ K +I+ +YVDD+
Sbjct: 58 FLLKRE*ELLIIQVYVDDI 2
Score = 41.2 bits (95), Expect(2) = 2e-38
Identities = 20/57 (35%), Positives = 32/57 (56%)
Frame = -3
Query: 272 EPEYFGEAMQGNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEE 328
EP+ EA++ D W +M++E+ +++ W L PEGK + RWV+R K E
Sbjct: 588 EPKNVKEALRDAD---WINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNKLE 427
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 155 bits (392), Expect = 6e-38
Identities = 77/183 (42%), Positives = 117/183 (63%), Gaps = 1/183 (0%)
Frame = -1
Query: 273 PEYFGEAMQGNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGS 332
P + EAM+ + W+ ++ E ++ KN TW ++LP+GKKA+ +RW++ +K ++DGS
Sbjct: 558 PRSYEEAMEDKE---WKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGS 388
Query: 333 -RRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHG 391
R K RLV +GF G D+ E F+PV K+ TIR++LS+ L Q+DVK AFL G
Sbjct: 387 IERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQG 208
Query: 392 DLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDH 451
+LE+E+YM P G E L + V +L K++YGLKQ+PR WY K + ++ GF + ++DH
Sbjct: 207 ELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDH 28
Query: 452 CCF 454
F
Sbjct: 27 TLF 19
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 146 bits (369), Expect = 3e-35
Identities = 82/220 (37%), Positives = 126/220 (57%), Gaps = 2/220 (0%)
Frame = +1
Query: 513 RSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYA 572
++E + + Q KYV LL+RF + +N P+ K K DE + Y
Sbjct: 13 QNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIK------DENGVKVDATKYK 174
Query: 573 SAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNN 632
VG LMY + TRPD+ + + ++SRFM+ P + H VK +LRYL G+ + + ++RN
Sbjct: 175 QIVGCLMY-LAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNG 351
Query: 633 LT-LQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAA 691
L+ ++D+D GD D KST+GY+F L AVSW SK Q V LSTT++E++A + A
Sbjct: 352 SEKLEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCA 531
Query: 692 KEMIWLKSFLKELGKEQDVP-PLFSDSQSVIFLAKNPVFH 730
+ +W++ L++LG Q ++ D+ S I L+KNPV H
Sbjct: 532 CQSVWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 146 bits (368), Expect = 3e-35
Identities = 89/249 (35%), Positives = 141/249 (55%), Gaps = 1/249 (0%)
Frame = +2
Query: 414 VCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALYVDDML 473
VC+L KS+YGLKQA RQWY K +E + + G+ + D F K S+ L +YVDD++
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 474 IAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRF 533
+AG++++EI +K + + F++KDLG + LG+ ++R++ +G+L L+Q KY +LL+
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSK-QGIL-LNQRKYTLELLEDS 373
Query: 534 NVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAV 593
+ TP LK SP ++E Y +G L+Y + TRPDI+ AV
Sbjct: 374 GNLAVKSTLTPYDISLKLHNSDSPLYNDE------TQYRRLIGKLIY-LTTTRPDISFAV 532
Query: 594 GVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCF-RRNNLTLQEFSDADLGGDSDGGKS 652
+S+F+S P + H++ +L+YLK + L + +NL L F+D+D KS
Sbjct: 533 QQLSQFVSKPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKS 712
Query: 653 TTGYIFTLG 661
TGY LG
Sbjct: 713 VTGYWVFLG 739
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 141 bits (356), Expect = 9e-34
Identities = 69/164 (42%), Positives = 103/164 (62%), Gaps = 3/164 (1%)
Frame = +1
Query: 609 ECVKWILRYLKGSSRMCLCFRR---NNLTLQEFSDADLGGDSDGGKSTTGYIFTLGGTAV 665
+ +KW+L+YL S + L + + L+ + DAD G+ D KS +G++FTL GT +
Sbjct: 1 QALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTI 180
Query: 666 SWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVIFLAK 725
SWK+ Q+ V LSTT++EY+A E K+ IWLK + ELG Q+ + DSQS I LA
Sbjct: 181 SWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEYVKIHCDSQSAIHLAN 360
Query: 726 NPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTK 769
+ V+H R KHI ++ HFIR++I +E+ + K+ ENP D+ TK
Sbjct: 361 HQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 135 bits (339), Expect(2) = 2e-32
Identities = 62/113 (54%), Positives = 77/113 (67%)
Frame = +1
Query: 48 TPEQNGVAERMNRTLNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEV 107
TP+QNGVAERMNRTL ER R M +G+ K FW +A+ TA Y+INR PS +D + P E+
Sbjct: 85 TPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPMEM 264
Query: 108 WSGKEVSLSHLKVFGCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWD 160
W GK V S L VFGC YV+ +S R KLDPK+ KC F+GY ++ GY WD
Sbjct: 265 WKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWD 423
Score = 23.1 bits (48), Expect(2) = 2e-32
Identities = 9/17 (52%), Positives = 10/17 (57%)
Frame = +3
Query: 23 GEYDSQEFKKFCSENGI 39
GEY EF FC + GI
Sbjct: 9 GEYVDGEFLAFCKQEGI 59
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 114 bits (284), Expect = 2e-25
Identities = 57/136 (41%), Positives = 88/136 (63%), Gaps = 1/136 (0%)
Frame = +2
Query: 641 ADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSF 700
+D GD++ KST+GY F LG A+SW SK Q VA ST E+EY+A + A + +WL+
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 701 LKELGKEQDVP-PLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILG 759
L+ + EQ+ P ++ D++S I L+KNPVFH R KHI +++H IRELI+++E+ +
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 760 SENPTDMLTKTVTADN 775
E D+ TK + ++
Sbjct: 362 EEKIADIFTKPLKIES 409
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 87.8 bits (216), Expect = 1e-17
Identities = 42/89 (47%), Positives = 62/89 (69%), Gaps = 1/89 (1%)
Frame = +2
Query: 304 TWSLTKLPEGKKALQNRWVYRLKEESDGSR-RYKARLVVKGFQQKQGIDFTEIFSPVVKM 362
T L K P G K + RW+Y++K DG+ +YKARLV KG+ ++QGIDF E+F+PVV++
Sbjct: 56 TLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVRI 235
Query: 363 TTIRVILSIVAAENLHLEQLDVKIAFLHG 391
TI ++L++ A + +DVKIAFL+G
Sbjct: 236 ETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 87.0 bits (214), Expect = 3e-17
Identities = 50/126 (39%), Positives = 74/126 (58%), Gaps = 4/126 (3%)
Frame = +1
Query: 583 VCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCF----RRNNLTLQEF 638
+C RPDI ++V V+S+FM +P K H ILRY++G+ L F + L +
Sbjct: 112 LC*RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICY 291
Query: 639 SDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLK 698
SD+D GD +ST+GY+F A+SW +K Q ALS+ E+EY+A + A + +WL
Sbjct: 292 SDSDWCGDR---RSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLD 462
Query: 699 SFLKEL 704
S +KEL
Sbjct: 463 SVIKEL 480
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 84.3 bits (207), Expect = 2e-16
Identities = 51/155 (32%), Positives = 83/155 (52%)
Frame = -2
Query: 87 AAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVSYVLIDSDRRDKLDPKAIKCFF 146
A YLINR P+ L Q P EV + ++ SL++++VFGC+ YVL+ + R+KL+ ++ K F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 147 IGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYKDRSSAESMSSSKQLKLSERVALEEI 206
IGY GY+ +D + ++++ SR+V F E Y + + E + + L + L I
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDL---RDLTSDKAGVLRVI 354
Query: 207 SESDVVKRNQINPENEDDEVEVELEQEPTTVIETP 241
E +K NQ ++ E EP +TP
Sbjct: 353 LEGLGIKMNQ--DQSTRSRQPEESSNEPRRAAQTP 255
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 81.3 bits (199), Expect = 1e-15
Identities = 44/83 (53%), Positives = 54/83 (65%), Gaps = 1/83 (1%)
Frame = -1
Query: 392 DLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDH 451
+LEE+IYMTQPEGF G ++ VCKL KSLYGLKQ+PRQWYK+F+ + S+ M
Sbjct: 249 ELEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSYRSSWATTGVLMTV 70
Query: 452 CCF-VKKFADSYIILALYVDDML 473
+ YI L LYVDDML
Sbjct: 69 VST*TR*RMSRYIYLVLYVDDML 1
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 78.2 bits (191), Expect = 1e-14
Identities = 44/122 (36%), Positives = 64/122 (52%), Gaps = 1/122 (0%)
Frame = +1
Query: 426 QAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFAD-SYIILALYVDDMLIAGSNMTEINR 484
Q+PR W+ +F + G+ +C DH F+K + IL +YVDD+ + G + I R
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 485 LKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTP 544
LK ++E FE+KDLG K LGM ++R + +SQ KYV LL + T P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGS--SISQRKYVLDLLKETRMIGCKTIRDP 354
Query: 545 LG 546
G
Sbjct: 355 YG 360
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 63.2 bits (152), Expect = 4e-10
Identities = 39/91 (42%), Positives = 50/91 (54%)
Frame = -3
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
V N KIK L+SDNGGEY S FK +GI + TP+QNGVA+R N+ L E
Sbjct: 269 VTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEV 90
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPS 96
AR + Q+ ++TA YLIN P+
Sbjct: 89 ARSLMFQAN---------VSTACYLINWIPT 24
>BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 514
Score = 61.6 bits (148), Expect = 1e-09
Identities = 39/113 (34%), Positives = 59/113 (51%)
Frame = -2
Query: 119 KVFGCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESV 178
++FGC S+V I SD R K D +A+KC FI Y GYR + ++K SR+VTF+E
Sbjct: 456 RIFGCTSFVHIHSDGRSKFDHRALKCVFIRYSSTQKGYRCYHPPSRKYFVSRDVTFHEQE 277
Query: 179 LYKDRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELE 231
Y ++ + SS + K E + L D+ +I E D VE +++
Sbjct: 276 SYFGQN*S---SSGGKYKEDESLLL-----LDLTFGPEIEVETGGDNVETDVD 142
>BF631997 weakly similar to GP|18542925|gb Putative pol polyprotein {Oryza
sativa}, partial (6%)
Length = 650
Score = 43.5 bits (101), Expect = 3e-04
Identities = 22/44 (50%), Positives = 27/44 (61%)
Frame = -2
Query: 18 KSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
K +NG E S EF + C E I T+ TP++NGVAERMN T
Sbjct: 550 KLNNGLEICSAEFNELCKEEHITRQYTVRNTPQKNGVAERMNIT 419
Score = 41.6 bits (96), Expect = 0.001
Identities = 48/161 (29%), Positives = 71/161 (43%), Gaps = 14/161 (8%)
Frame = -1
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFC----SENGIRMIKTILG----TPEQNG 53
WK VE+QT K+K L+++ + FC + G K I G T E
Sbjct: 599 WKILVESQTGKKVKRLQTE*------RLGNLFCRVQ*TLQGRTHYKAIYGEKYSTKEWCC 438
Query: 54 VAERMNRTLNERARCMRIQSGLPKMFWVDAINTAAYLINRGP------SIPLDYQLPEEV 107
A N ERARCM +GL + F +AI+T YL+N P + L Y L
Sbjct: 437 *ANEYN--FLERARCMFSNAGLNRSFQAEAISTKCYLVNVLPLLL*TVRLHLRYGL---- 276
Query: 108 WSGKEVSLSHLKVFGCVSYVLIDSDRRDKLDPKAIKCFFIG 148
++L ++ GC +Y ++ KL+P++ K F G
Sbjct: 275 -----INLLITQILGCPAYYHVN---EGKLEPRSKKGLFWG 177
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,119,248
Number of Sequences: 36976
Number of extensions: 310915
Number of successful extensions: 1614
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 1584
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1592
length of query: 799
length of database: 9,014,727
effective HSP length: 104
effective length of query: 695
effective length of database: 5,169,223
effective search space: 3592609985
effective search space used: 3592609985
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0065.18


