

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.12
(1309 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
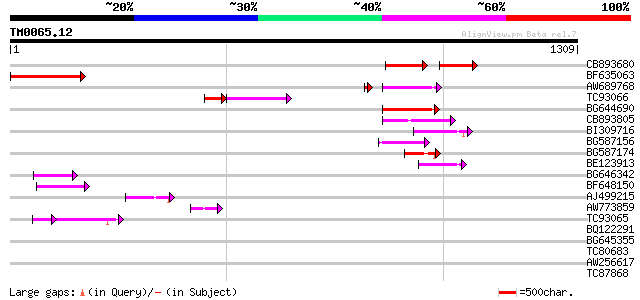
Score E
Sequences producing significant alignments: (bits) Value
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 84 7e-32
BF635063 weakly similar to PIR|F84486|F84 probable retroelement ... 126 5e-29
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 103 6e-23
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 61 3e-20
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 92 2e-18
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 84 3e-16
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 80 4e-15
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 74 3e-13
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 74 4e-13
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 67 5e-11
BG646342 weakly similar to PIR|F84486|F84 probable retroelement ... 60 4e-09
BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x pa... 51 3e-06
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 50 4e-06
AW773859 46 1e-04
TC93065 34 3e-04
BQ122291 43 0.001
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 42 0.002
TC80683 homologue to GP|8777424|dbj|BAA97014.1 gb|AAF56406.1~gen... 41 0.003
AW256617 39 0.017
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 38 0.029
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 83.6 bits (205), Expect(2) = 7e-32
Identities = 46/98 (46%), Positives = 66/98 (66%)
Frame = -2
Query: 867 RGLIMMRFFSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSE 926
+G I++ F P+V+ +I +L++VA +++LE +DVKT FL G+L E IYM QPEGFS
Sbjct: 578 KGRILL*NFVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS* 399
Query: 927 TGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYR 964
G++V KLK+S+YGLKQ PRQ + R G+R
Sbjct: 398 E-VGKMVGKLKKSMYGLKQGPRQCI*SLKALCTRKGFR 288
Score = 73.6 bits (179), Expect(2) = 7e-32
Identities = 40/89 (44%), Positives = 55/89 (60%)
Frame = -3
Query: 992 MLIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLSQKSYVEGVLS 1051
+L+ +++ + LKT+ KE DMKDLG AKKI+GM+I D+ L LSQ Y+ VL
Sbjct: 271 LLVVGSNIDEIKNLKTRFSKEIDMKDLGPAKKIIGMQIMIDKQKGVL*LSQVEYITRVLQ 92
Query: 1052 RFDMSKENPVSTPLANHFKLSLGQCSKTD 1080
F+M VST LA+HF LS Q +T+
Sbjct: 91 IFNMGNAILVSTTLASHFCLSHEQSPQTE 5
>BF635063 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 677
Score = 126 bits (317), Expect = 5e-29
Identities = 66/178 (37%), Positives = 110/178 (61%), Gaps = 3/178 (1%)
Frame = -2
Query: 1 GAKFEVTRFYGTGNFGLWHRRVKDLLAQQSLQKALRDEK--PTDIATVDWNEMKEKAASL 58
G+K ++ +F G +FGLW +++ +L QQ +KAL+ E P ++ + EM +KA S
Sbjct: 565 GSKRDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEVSLPVTMSRAEKTEMVDKARSA 386
Query: 59 IRLCVSDDLMNHILDLTTPKDVWDKLESLYMSKTLMNKLFAKQRLYSLKMQEGGDLQAHV 118
I LC+ D ++ + T +W KL SLYM+K+L ++ F KQ+LYS +M E + +
Sbjct: 385 IVLCLGDKVLREVAKERTAASMWAKL*SLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQL 206
Query: 119 YAFNNILADLTRLRVKVDDEDKAIILLRSLPGSYDHLVTTLTYGKD-SIKLDSISSTL 175
FN IL DL + V+++DE+KAI+LL +LP S++ T+ YGK+ ++ L+ + + L
Sbjct: 205 TEFNKILDDLENIEVQLEDEEKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAAL 32
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 103 bits (258), Expect(2) = 6e-23
Identities = 54/139 (38%), Positives = 84/139 (59%), Gaps = 2/139 (1%)
Frame = +1
Query: 861 KGIHSKRGLIMMRFFSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQ 920
KG G FSPV++ +IR +L + + ++Q+D+ FL+G L+E++YM Q
Sbjct: 262 KGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQ 441
Query: 921 PEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGY--RRCDYDCCVYVRSLD 978
P+GF E + LVCKL +SLYGLKQ+PR Y+ S ++ G+ RCD +Y +
Sbjct: 442 PQGF-EAANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQ--- 609
Query: 979 DGSFIFLLLYVDDMLIAAN 997
+G+ I+L +YVDD+LI +
Sbjct: 610 NGACIYLXIYVDDILITGS 666
Score = 23.1 bits (48), Expect(2) = 6e-23
Identities = 11/19 (57%), Positives = 13/19 (67%)
Frame = +2
Query: 820 RHGILFSCHMERELLVASG 838
+HGILF R+LL ASG
Sbjct: 143 KHGILFLFLHTRKLLAASG 199
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 61.2 bits (147), Expect(2) = 3e-20
Identities = 26/52 (50%), Positives = 37/52 (71%), Gaps = 1/52 (1%)
Frame = +1
Query: 450 CKYCVL-GKQCRVRFKTGQHKTKGILDYVHSDVWGPTKEPSVGGFRYFLHLL 500
CK+ + G + +V F T H+TKGILDY+HSD+WGP+K S GG RY + ++
Sbjct: 22 CKHLLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTII 177
Score = 56.6 bits (135), Expect(2) = 3e-20
Identities = 53/152 (34%), Positives = 72/152 (46%), Gaps = 2/152 (1%)
Frame = +2
Query: 500 LMIFLVRSGFIF*NINLKSLPSSNCGRQRLRIRQGEK*SICGLIMELNTLTRTSCVSVRR 559
LMIFL R GFIF I ++ P S G L++RQG * I++ +++ T S +
Sbjct: 176 LMIFLGRFGFIFCGIKMRLFPHSRSGEFLLKLRQGRM*RSS*QIID*SSVVVTLMSSAQI 355
Query: 560 MEFRDTFL*GRHHSRMVLQKG*TRL*QRRQGAFG*MLVFQ--KVSGQQQSIWRAI*STGH 617
M DT ++ VLQ *+ L R ML ++ + G +Q + ST
Sbjct: 356 MVLLDTKPFQGIPNKTVLQNE*SGLYLRELDVCSQMLGYRIDVIFGSRQHLLHVTWSTVL 535
Query: 618 HELLWMEKLQNRCGQVTPLISVI*GFLGVQHM 649
H K Q GQV LI +I* FL VQHM
Sbjct: 536 HIQHLTSKFQKIFGQVILLIILI*EFLDVQHM 631
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 91.7 bits (226), Expect = 2e-18
Identities = 46/132 (34%), Positives = 81/132 (60%)
Frame = -2
Query: 861 KGIHSKRGLIMMRFFSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQ 920
+G + K G+ FSPV R +IR ++A A L QMDVK+ F++G+L+E+++++Q
Sbjct: 394 QGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFINGDLKEEVFVKQ 215
Query: 921 PEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDG 980
P GF + V +L ++LYGLKQ+PR Y+R ++L+ G++R D +++ +
Sbjct: 214 PPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDNTLFLLK-RE* 38
Query: 981 SFIFLLLYVDDM 992
+ + +YVDD+
Sbjct: 37 ELLIIQVYVDDI 2
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 84.3 bits (207), Expect = 3e-16
Identities = 53/173 (30%), Positives = 91/173 (51%), Gaps = 4/173 (2%)
Frame = +3
Query: 861 KGIHSKRGLIMMRFFSPVVRHTSIRAVLALVAS--RD-MHLEQMDVKTTFLHGNLEEQIY 917
KG + G+ F+PV R +IR V+AL A RD + + M + + + + +
Sbjct: 243 KGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KAHSCMEN*MRKFLL 422
Query: 918 MEQPEGFSETGDGRLVC-KLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRS 976
+ R++ ++KR+LYGLKQ+PR Y R ++Y + G+ +C Y+ ++V+
Sbjct: 423 INH-----RVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKCPYEHTLFVKL 587
Query: 977 LDDGSFIFLLLYVDDMLIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEI 1029
+ G + + LYVDD++ N + E K + KEF+M DLG LG+E+
Sbjct: 588 SEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLGVEV 746
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 80.5 bits (197), Expect = 4e-15
Identities = 48/148 (32%), Positives = 83/148 (55%), Gaps = 13/148 (8%)
Frame = +2
Query: 933 VCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDM 992
VC+L++S+YGLKQ+ RQ Y + ++ GY + D ++ + D SF LL+YVDD+
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTK-FKDSSFTTLLVYVDDI 196
Query: 993 LIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLSQKSY------- 1045
++A N + + +K L F +KDLG+ + LG+E+ R + + + L+Q+ Y
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSK--QGILLNQRKYTLELLED 370
Query: 1046 -----VEGVLSRFDMS-KENPVSTPLAN 1067
V+ L+ +D+S K + +PL N
Sbjct: 371 SGNLAVKSTLTPYDISLKLHNSDSPLYN 454
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 74.3 bits (181), Expect = 3e-13
Identities = 43/118 (36%), Positives = 65/118 (54%)
Frame = -1
Query: 852 KSSRLA*LQKGIHSKRGLIMMRFFSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGN 911
K +RL + +G G + F+PV + +IR VL+L + L QMDVK FL G
Sbjct: 378 KKTRL--VARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGE 205
Query: 912 LEEQIYMEQPEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYD 969
LE+++YM P G V +LK+++YGLKQSPR Y + + + G+R+ + D
Sbjct: 204 LEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELD 31
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 73.9 bits (180), Expect = 4e-13
Identities = 44/89 (49%), Positives = 54/89 (60%), Gaps = 7/89 (7%)
Frame = -1
Query: 912 LEEQIYMEQPEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCC 971
LEE+IYM QPEGF G VCKL++SLYGLKQSPRQ YKRFDSY R +
Sbjct: 246 LEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSY-------RSSWATT 88
Query: 972 VYVRSLDD-------GSFIFLLLYVDDML 993
+ ++ +I+L+LYVDDML
Sbjct: 87 GVLMTVVST*TR*RMSRYIYLVLYVDDML 1
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 67.0 bits (162), Expect = 5e-11
Identities = 38/111 (34%), Positives = 60/111 (53%)
Frame = +1
Query: 945 QSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLIAANHLHNVNE 1004
QSPR + RF + + GY +C D ++++ L++YVDD+ + +H +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 1005 LKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLSQKSYVEGVLSRFDM 1055
LK L +EF++KDLG K LGME+ R + K +SQ+ YV +L M
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWK--KGSSISQRKYVLDLLKETRM 327
>BG646342 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 599
Score = 60.5 bits (145), Expect = 4e-09
Identities = 35/103 (33%), Positives = 53/103 (50%), Gaps = 2/103 (1%)
Frame = +2
Query: 55 AASLIRLCVSDDLMNHILDLTTPKDVWDKLESLYMSKTLMNKLFAKQRLYSLKMQEGGDL 114
A S I LC+ D ++ + T + KLE LYM+K+L ++ F KQ+LYS KM E +
Sbjct: 2 ARSAIVLCLGDKVLREVAKEPTATSMCAKLEYLYMTKSLAHRQFLKQQLYSFKMVESKAI 181
Query: 115 QAHVYAFNNILADLTRLRVKVDDEDKAII--LLRSLPGSYDHL 155
+ FN I+ DL + V ++D ++ L G HL
Sbjct: 182 TELLVEFNKIIGDLENIEVHLEDAGALMVWCCLEDEEGDVSHL 310
>BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x paradisi},
partial (3%)
Length = 658
Score = 51.2 bits (121), Expect = 3e-06
Identities = 31/122 (25%), Positives = 57/122 (46%), Gaps = 1/122 (0%)
Frame = +2
Query: 63 VSDDLMNHILDLTTPKDVWDKLESLYMSKTLMNKLFAKQRLYSLKMQEGGDLQAHVYAFN 122
+SD L + + KD+WDKLE+ YM + +K F + KM + + +Y
Sbjct: 224 MSDSLFDIYQSSPSAKDLWDKLETRYMREDATSKKFLVSHFNNYKMVDNKSVMEQLYEIE 403
Query: 123 NILADLTRLRVKVDDEDKAIILLRSLPGSYDHLVTTLTYGKDSIKLDSISSTL-LQHAQR 181
IL + + + +D+ ++ LP S+ T+ + K+ I L+ + + L L R
Sbjct: 404 RILNNYKQHNMNMDETIIVSSIIDKLPPSWKDFKRTMKHKKEDISLEQLGNHLRLXEEYR 583
Query: 182 RQ 183
+Q
Sbjct: 584 KQ 589
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 50.4 bits (119), Expect = 4e-06
Identities = 32/126 (25%), Positives = 59/126 (46%), Gaps = 12/126 (9%)
Frame = +3
Query: 267 SEEDLLCVSSVKCTDA---WVLDSGCSYHMTPHREWFNSFKLGDFGYVYLGDDKPCIIKG 323
S+E LL +S+ W++DSGC++HMT R+ F V + + ++G
Sbjct: 51 SKEQLLAMSTFATKQPSKYWLIDSGCTHHMTHDRDLFKELNKSTISKVRMLNGAHIEVEG 230
Query: 324 MRQVKIALDDGRVRTLSQVRYVPEVTKNMISLGTLHENGYS---------FKSEENRDIL 374
+ V + G + +S V Y P++ ++++S+ L GY K + N+++L
Sbjct: 231 IGTVLVKSHSG-YKQISNVLYAPKLNQSLLSVPQLLTKGYKVLFEHEKCVIKDQNNKEVL 407
Query: 375 RVSKGA 380
R A
Sbjct: 408 RAQMKA 425
>AW773859
Length = 538
Score = 45.8 bits (107), Expect = 1e-04
Identities = 20/74 (27%), Positives = 35/74 (47%)
Frame = -3
Query: 417 MWHMRLGHLSERGMMQLHKRNLLKGVRSCTIGLCKYCVLGKQCRVRFKTGQHKTKGILDY 476
+WH RLGHLS R ++ LH + ++ C C + ++ F+ ++ +
Sbjct: 230 LWHFRLGHLSNRKLLSLHSNFPFITIDQNSV--CDICHYSRHKKLPFQLSTNRASKCYEL 57
Query: 477 VHSDVWGPTKEPSV 490
H D+WGP S+
Sbjct: 56 FHFDIWGPFSTQSI 15
>TC93065
Length = 783
Score = 34.3 bits (77), Expect(2) = 3e-04
Identities = 35/171 (20%), Positives = 73/171 (42%), Gaps = 9/171 (5%)
Frame = +2
Query: 100 KQRLYSLKMQEGGDLQAHVYAFNNILADLTRLRVKVDDEDKAIILLRSLPGSYDHLVTTL 159
++ +LKM+E ++ + ++ + L + D+ +L LP ++ +++L
Sbjct: 218 RREFEALKMKETETVREFSDRISKVVTQIRLLGEDLSDQRVVEKILVCLPEMFEAKISSL 397
Query: 160 TYGKDSIKLDSISST-LLQHAQRRQSVEEGGGSSGKCLFVKGGQG---RARGKGKAVDSG 215
K+ ++ LQ +++R+S+ G L G+ ++ GK K
Sbjct: 398 EENKNFSEITVAELVNALQASEQRRSLRMEENVEGAFLANNKGKNQSFKSFGKKKFPPCP 577
Query: 216 KKKRSKSKD-----RKTAECYSCKQIGHWKRDCPNRPGKSSNSNSSANVVQ 261
K+ D R +C +C Q+GH ++ C N K++ A VV+
Sbjct: 578 HCKKDTHLDKFCWYRPGVKCRACNQLGHVEKVCKN---KTNQQEQEARVVE 721
Score = 29.3 bits (64), Expect(2) = 3e-04
Identities = 16/54 (29%), Positives = 24/54 (43%)
Frame = +3
Query: 54 KAASLIRLCVSDDLMNHILDLTTPKDVWDKLESLYMSKTLMNKLFAKQRLYSLK 107
+A ++I + DD+ IL+L K+ W KL Y K+ SLK
Sbjct: 72 RALAIIHAALHDDIFIKILNLEIAKEAWSKLMEEYQGSERTKKMKVLNLEESLK 233
>BQ122291
Length = 487
Score = 42.7 bits (99), Expect = 0.001
Identities = 33/156 (21%), Positives = 68/156 (43%), Gaps = 2/156 (1%)
Frame = -2
Query: 34 ALRDEKPTDIATVDWNEMKEKAASLIRLCVSDDLMNHILDLTTPKDVWDKLESLYMS--K 91
A R+ + A +W + + I +S L++ + L VWD++ S + K
Sbjct: 483 AAREAGTRNPAYTEWEDKDSLLCTWILSRISPSLLSRFVLLRHSWQVWDEIHSYCFTQMK 304
Query: 92 TLMNKLFAKQRLYSLKMQEGGDLQAHVYAFNNILADLTRLRVKVDDEDKAIILLRSLPGS 151
T +L ++ R + + + A + A + LA + V D ++L +LP
Sbjct: 303 TRSRQLRSELRSITKGSRTVSEFIARIRAISESLASIGD---PVSHRDLIEVVLEALPEE 133
Query: 152 YDHLVTTLTYGKDSIKLDSISSTLLQHAQRRQSVEE 187
+D +V ++ + + LD + S LL R++ ++
Sbjct: 132 FDPIVASVNAKSEVVSLDELESQLLTQESRKEKFKK 25
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 42.0 bits (97), Expect = 0.002
Identities = 29/91 (31%), Positives = 35/91 (37%), Gaps = 8/91 (8%)
Frame = -1
Query: 184 SVEEGGGSSGKCLFVKGGQGRARGKGKAVDSGKKKRSKSKDRKTAECYSCKQIGHWKRDC 243
+V GG+SGKC K Q S + S + CY C Q GHW +C
Sbjct: 534 TVSGSGGASGKCY--KCQQPGHWASNCPSMSAANRVSGGSGGASGNCYKCNQPGHWANNC 361
Query: 244 PN---RPGKSSNSNSSANVV-----QSDGSC 266
PN P NSN+ Q DG C
Sbjct: 360 PNMSAAPQSHGNSNTGQGRYGIAQNQPDGRC 268
Score = 34.3 bits (77), Expect = 0.32
Identities = 21/76 (27%), Positives = 34/76 (44%)
Frame = -1
Query: 191 SSGKCLFVKGGQGRARGKGKAVDSGKKKRSKSKDRKTAECYSCKQIGHWKRDCPNRPGKS 250
SS +CL ++ + G G V++ S +CY C+Q GHW +CP+ +
Sbjct: 600 SSAQCLHLRNPPEQTAG-GAYVNTVSGSGGAS-----GKCYKCQQPGHWASNCPSMSAAN 439
Query: 251 SNSNSSANVVQSDGSC 266
S S + G+C
Sbjct: 438 RVSGGSGG---ASGNC 400
>TC80683 homologue to GP|8777424|dbj|BAA97014.1
gb|AAF56406.1~gene_id:K9P8.7~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (16%)
Length = 1360
Score = 41.2 bits (95), Expect = 0.003
Identities = 33/134 (24%), Positives = 57/134 (41%), Gaps = 33/134 (24%)
Frame = +1
Query: 162 GKDSIKLDSISSTLLQHAQRRQSVEEGGGSSGKCLFVKGGQGRARG-KGKAVDSGKKKRS 220
GKD KL IS L HA+ ++++ GGG +G +G+ + K K D +++RS
Sbjct: 793 GKDKSKLKDISGDL--HAKDAENLKTGGGKK----ISRGQKGKLKKMKEKYADQDEEERS 954
Query: 221 ------------------------KSKDRKTAE--------CYSCKQIGHWKRDCPNRPG 248
K +K+ CY CK++GH RDC +P
Sbjct: 955 IRMSLLASSGKPIKKEETLPVIETSDKGKKSDSGPIDAPKICYKCKKVGHLSRDCKEQPN 1134
Query: 249 KSSNSNSSANVVQS 262
+S++++ ++
Sbjct: 1135DLLHSHATSEAEEN 1176
>AW256617
Length = 718
Score = 38.5 bits (88), Expect = 0.017
Identities = 24/72 (33%), Positives = 39/72 (53%)
Frame = -2
Query: 346 PEVTKNMISLGTLHENGYSFKSEENRDILRVSKGAMTVMRAKRTAGNIYKLLGGTVMGDV 405
P + K S+ TL NG++ + ++DILR+ GA LLG T++ D+
Sbjct: 330 P*IKK*FDSMHTL*ANGFANRINRDKDILRLRNGA---------------LLGKTIV*DI 196
Query: 406 ASVETDDDATKM 417
ASV+ D+DA ++
Sbjct: 195 ASVDPDNDAAEI 160
Score = 26.2 bits (56), Expect(2) = 5.3
Identities = 12/37 (32%), Positives = 20/37 (53%)
Frame = -2
Query: 199 KGGQGRARGKGKAVDSGKKKRSKSKDRKTAECYSCKQ 235
KG R +G+ K+ ++K RK +C++CKQ
Sbjct: 564 KGRVSRTC*EGRQTSFCKEIEVENKSRKIFQCHNCKQ 454
Score = 22.3 bits (46), Expect(2) = 5.3
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = -1
Query: 159 LTYGKDSIKLDSISSTLLQHAQ 180
L GKD+IKL+ I++ + Q
Sbjct: 643 LINGKDTIKLEDIAAAITSRCQ 578
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 37.7 bits (86), Expect = 0.029
Identities = 24/69 (34%), Positives = 28/69 (39%)
Frame = +3
Query: 188 GGGSSGKCLFVKGGQGRARGKGKAVDSGKKKRSKSKDRKTAECYSCKQIGHWKRDCPNRP 247
GGG G GG GR RG G S K CY C + GH+ R+C NR
Sbjct: 264 GGGGGG------GGGGRGRGGGGGGGSDLK------------CYECGEPGHFARECRNRG 389
Query: 248 GKSSNSNSS 256
G + S
Sbjct: 390 GGGAGRRRS 416
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.337 0.146 0.474
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,465,189
Number of Sequences: 36976
Number of extensions: 664217
Number of successful extensions: 5001
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 2715
Number of HSP's successfully gapped in prelim test: 246
Number of HSP's that attempted gapping in prelim test: 2110
Number of HSP's gapped (non-prelim): 3190
length of query: 1309
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1201
effective length of database: 5,021,319
effective search space: 6030604119
effective search space used: 6030604119
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0065.12


