

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
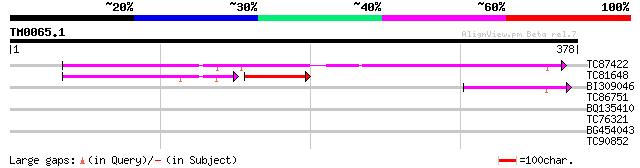
Query= TM0065.1
(378 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87422 similar to PIR|T00840|T00840 probable senescence-related... 201 4e-52
TC81648 similar to PIR|T00840|T00840 probable senescence-related... 72 9e-20
BI309046 similar to GP|15320412|dbj ERD7 protein {Arabidopsis th... 60 2e-09
TC86751 similar to GP|18252881|gb|AAL62367.1 ubiquitin activatin... 28 5.8
BQ135410 28 5.8
TC76321 homologue to GP|16033631|gb|AAL13304.1 leucine zipper-co... 28 7.6
BG454043 similar to GP|2213425|emb| hypothetical protein {Citrus... 28 7.6
TC90852 27 9.9
>TC87422 similar to PIR|T00840|T00840 probable senescence-related protein
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1660
Score = 201 bits (511), Expect = 4e-52
Identities = 121/358 (33%), Positives = 195/358 (53%), Gaps = 22/358 (6%)
Frame = +2
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+ VL+ +P ++L+++ +V LA G I+ + ++ +A + ++G+ +QWPL KD
Sbjct: 185 EHVLVTVPNVILHLIEKDSSVHLASGDLTIVSLKEEEKVVAVLARIGDQIQWPLAKDVST 364
Query: 96 VKVDTLHYLFTLPVKDGGSEPLSYGVTFPEQCYGSMGLLESF---LKDHSCFSDLKT-GK 151
VK+D HY FTL + E L+YG+T + GS +L+ L+ +S S K G
Sbjct: 365 VKLDESHYFFTLKLPQSDDEVLNYGLTVAAK--GSKKVLKKLDEVLEKYSLLSVEKVKGV 538
Query: 152 KG---------------GLDWEQFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQ 196
KG W AP VEDY+ + IA G+GQ+++GI C + ++
Sbjct: 539 KGWEVLEKKEDVEENSAAAYWTTLAPNVEDYSGRFGRWIAAGSGQMIRGILWCGDVTVDR 718
Query: 197 VQSGGQTILTDNKNNGLVRQSVSNNKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDG 256
++ G + +++ + + A+N ++K V++LT M+EK++ +L G
Sbjct: 719 LKWG----------DDFMKKRLQPGSSQSQISPLALN-SMKSVKKLTKMSEKVALGVLSG 865
Query: 257 VGIMSGSVMAPVLKSQPGQAFLKMLPGEVLLASLDAVNKVFEATEAAQKQSLSATSQAAT 316
+SG + + V+ S+PG+ F LPGEVLLASLD N+V +A E A + +S +S T
Sbjct: 866 AVKVSGFLTSSVVNSKPGKKFFSFLPGEVLLASLDGFNRVCDAVEVAGRNVMSTSSVVTT 1045
Query: 317 RMVSNRYGEEAGEATEHVFATAGHAANTAWNVSKIRKAINP---VTPASTAGGALRNS 371
+VS++YGE+A E T F AGHA TAW V K+RKA+NP + P + A A+R S
Sbjct: 1046GLVSHKYGEKAAEITNEGFDAAGHAIGTAWAVFKLRKALNPKSVIKPTTLAKAAVRAS 1219
>TC81648 similar to PIR|T00840|T00840 probable senescence-related protein
[imported] - Arabidopsis thaliana, partial (36%)
Length = 854
Score = 72.0 bits (175), Expect(2) = 9e-20
Identities = 45/136 (33%), Positives = 70/136 (51%), Gaps = 19/136 (13%)
Frame = +1
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+++LI+IPG + L+D+ +VELA G F ++++ S+A ++ + +QWPL KDE
Sbjct: 211 EQILIKIPGAILNLIDQQYSVELASGDFTVVRLRQGENSIAVYARIADEIQWPLAKDETA 390
Query: 96 VKVDTLHYLFTLPVKDG----------------GSEPLSYGVTFPEQCYGSMGLLES--- 136
VKVD HY F+ G S+ LSYG+T + G LL+
Sbjct: 391 VKVDDSHYFFSFSAPKGYDSDEDEADRSKNSKTESDLLSYGLTIASK--GQEHLLKELDV 564
Query: 137 FLKDHSCFSDLKTGKK 152
L++ S FS K +K
Sbjct: 565 ILENCSNFSVQKVSEK 612
Score = 42.4 bits (98), Expect(2) = 9e-20
Identities = 17/44 (38%), Positives = 27/44 (60%)
Frame = +3
Query: 157 WEQFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSG 200
W AP VE+Y+ A+ IA G+G ++KGI C + +++Q G
Sbjct: 723 WTTLAPNVEEYSGTAARMIASGSGHVIKGILWCGDVTMDRLQWG 854
>BI309046 similar to GP|15320412|dbj ERD7 protein {Arabidopsis thaliana},
partial (18%)
Length = 439
Score = 59.7 bits (143), Expect = 2e-09
Identities = 34/78 (43%), Positives = 42/78 (53%), Gaps = 6/78 (7%)
Frame = -1
Query: 303 AQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAWNVSKIRKAIN------ 356
A K +S +S T +V +RYGEEA AT AGHA TAW KIRKAIN
Sbjct: 439 AGKNVMSTSSTVTTELVDHRYGEEAAHATNEGLNAAGHALGTAWAAFKIRKAINPKSVFK 260
Query: 357 PVTPASTAGGALRNSIKN 374
P T A +A A + +K+
Sbjct: 259 PTTLAKSAAKAAASDLKS 206
>TC86751 similar to GP|18252881|gb|AAL62367.1 ubiquitin activating enzyme -
like protein {Arabidopsis thaliana}, partial (98%)
Length = 1432
Score = 28.1 bits (61), Expect = 5.8
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 2/33 (6%)
Frame = -2
Query: 101 LHYLF--TLPVKDGGSEPLSYGVTFPEQCYGSM 131
+ Y+F TLP++ E + YG+ FP C+ ++
Sbjct: 1095 IRYVFYDTLPIRSIKIEKVFYGIPFPRNCFDNL 997
>BQ135410
Length = 782
Score = 28.1 bits (61), Expect = 5.8
Identities = 17/47 (36%), Positives = 24/47 (50%), Gaps = 3/47 (6%)
Frame = -1
Query: 84 GVQWPLTKDEPVVKVDTLHYLFTLPVKDGGSEPL---SYGVTFPEQC 127
G++W + E VD++H +L VK GSE L G FP +C
Sbjct: 713 GLEWRIFLQERFQSVDSIHQEGSLFVKVSGSESLWQ*VRGQFFPPRC 573
>TC76321 homologue to GP|16033631|gb|AAL13304.1 leucine zipper-containing
protein {Euphorbia esula}, partial (92%)
Length = 1468
Score = 27.7 bits (60), Expect = 7.6
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = +1
Query: 308 LSATSQAATRMVSNRYGEEAGEATEHVFA 336
LSATS AAT+ N Y +E + H+F+
Sbjct: 1315 LSATSTAATKQCGNIYHDEILVSLNHIFS 1401
>BG454043 similar to GP|2213425|emb| hypothetical protein {Citrus x
paradisi}, partial (18%)
Length = 663
Score = 27.7 bits (60), Expect = 7.6
Identities = 13/28 (46%), Positives = 17/28 (60%)
Frame = +2
Query: 308 LSATSQAATRMVSNRYGEEAGEATEHVF 335
LSATS AAT+ N Y +E + H+F
Sbjct: 578 LSATSTAATKQCGNIYHDEILXSLNHIF 661
>TC90852
Length = 639
Score = 27.3 bits (59), Expect = 9.9
Identities = 10/21 (47%), Positives = 14/21 (66%)
Frame = +3
Query: 87 WPLTKDEPVVKVDTLHYLFTL 107
WPL +D+ V K + +LFTL
Sbjct: 126 WPLDRDQTVQKANRFRFLFTL 188
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.130 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,141,951
Number of Sequences: 36976
Number of extensions: 103933
Number of successful extensions: 364
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 360
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 362
length of query: 378
length of database: 9,014,727
effective HSP length: 98
effective length of query: 280
effective length of database: 5,391,079
effective search space: 1509502120
effective search space used: 1509502120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0065.1


