

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
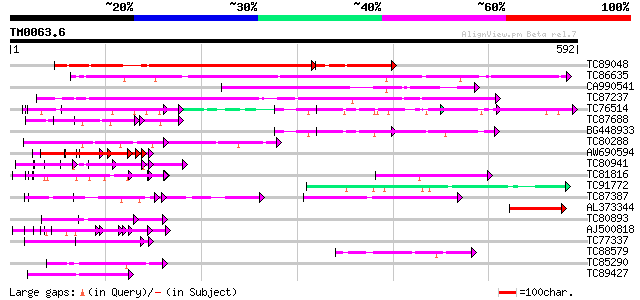
Query= TM0063.6
(592 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89048 weakly similar to PIR|T46215|T46215 hypothetical protein... 320 e-110
TC86635 weakly similar to GP|9758600|dbj|BAB09233.1 gene_id:MDF2... 330 9e-91
CA990541 weakly similar to GP|9758600|dbj gene_id:MDF20.10~pir||... 134 1e-31
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 93 3e-19
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 74 1e-13
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 69 5e-12
BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfa... 67 3e-11
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 66 3e-11
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 65 7e-11
TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein ... 65 7e-11
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 65 7e-11
TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown prot... 62 5e-10
TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I... 61 1e-09
AL373344 similar to GP|9758600|dbj| gene_id:MDF20.10~pir||T08929... 61 1e-09
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 60 2e-09
AJ500818 weakly similar to GP|10435304|dbj unnamed protein produ... 60 2e-09
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 60 3e-09
TC88579 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 58 9e-09
TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsi... 57 2e-08
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 56 3e-08
>TC89048 weakly similar to PIR|T46215|T46215 hypothetical protein T8P19.220
- Arabidopsis thaliana, partial (35%)
Length = 1329
Score = 320 bits (821), Expect(2) = e-110
Identities = 170/274 (62%), Positives = 205/274 (74%)
Frame = +2
Query: 47 EDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEG 106
+D + V++EA++ E K K +V E++ E+DE K E+ G E+ D++DED G E
Sbjct: 323 DDNKPKQVENEAKD-EPKPKEEVEVTEQDAEVAESDE-KKEIDGHEDSDKDDEDEGGDED 496
Query: 107 EGEGEEEEEQEEEEEAEAEADEKVKAKRGPKKGGKKETASEKKDPATPASERPTRERKTV 166
+ + +EEE+ EE KKG KKE+ SEKK P TP S+RPTRERKTV
Sbjct: 497 DADDDEEEDAGEE-----------------KKGVKKES-SEKKSPVTPTSDRPTRERKTV 622
Query: 167 ERYSVPSPSKSAKSSGGKGLVIEKGRGEQLKDIPNVAFKLSKRKPDDNLHALHSILFGKR 226
ERYS PSPSK +SS KGL+IEKGRG QLKDIPNVAFKLSKRKPDDNLH LHS+LFGK+
Sbjct: 623 ERYSEPSPSKFGRSSSSKGLIIEKGRGTQLKDIPNVAFKLSKRKPDDNLHMLHSLLFGKK 802
Query: 227 TKAYNLKRNIGQFSGFVWTENEEKQRAKVKERIDKYVKEKLVDFCDVLNIPVSKGNIKKE 286
TK NLKRNIGQFSG+VW ENEEKQR +VKERIDK VKEKL+ FCDVLNIP++K ++KKE
Sbjct: 803 TKVNNLKRNIGQFSGYVWAENEEKQRTRVKERIDKCVKEKLIVFCDVLNIPINKSSVKKE 982
Query: 287 ELSVKLLEFLENPQATTDVLLADNEKKVKKRTRK 320
ELS KLLEFLE+P ATTDVLLA+ E+K KKR +K
Sbjct: 983 ELSAKLLEFLESPHATTDVLLAEKEQKGKKRVKK 1084
Score = 97.1 bits (240), Expect(2) = e-110
Identities = 57/87 (65%), Positives = 66/87 (75%), Gaps = 2/87 (2%)
Frame = +1
Query: 320 KVTPSKSPGEASTETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQ--ENVAAP 377
K T SK+PGE TPAKK+K+TSESGKKRK SDVEED++AELSDAKDESQ E+VA
Sbjct: 1081 KATSSKTPGE----TPAKKKKQTSESGKKRK-PSDVEEDDQAELSDAKDESQEDEDVAVA 1245
Query: 378 HKESDDDESKSEEEEDKSKAQKKTSKK 404
+ SD++ KSE EED KA K TSKK
Sbjct: 1246 NNGSDNEVGKSEGEEDTPKAHKSTSKK 1326
Score = 40.4 bits (93), Expect = 0.002
Identities = 36/156 (23%), Positives = 65/156 (41%), Gaps = 2/156 (1%)
Frame = +2
Query: 333 ETPAKKQKKTSESGKKRKGSSDV-EEDEKAELSDAKDESQENVAAPHKESDDDESKSEEE 391
E AK + K E + + ++V E DEK E+ +D +++ + D+D++ +EE
Sbjct: 347 ENEAKDEPKPKEEVEVTEQDAEVAESDEKKEIDGHEDSDKDD---EDEGGDEDDADDDEE 517
Query: 392 EDKSKAQKKTSKKIVKEGSGSKAGERTKSAKKTPVKSSKNVDKTPKKSTPKQTAAEHDSA 451
ED + +K K+ S KK+PV + + +K+ + +
Sbjct: 518 EDAGEEKKGVKKE--------------SSEKKSPVTPTSDRPTRERKTVERYSEPSPSKF 655
Query: 452 SASLSKSKQPASKKRKTE-NEKPDTKGKASSKKQTD 486
S S K R T+ + P+ K S +K D
Sbjct: 656 GRSSSSKGLIIEKGRGTQLKDIPNVAFKLSKRKPDD 763
Score = 31.6 bits (70), Expect = 0.89
Identities = 15/27 (55%), Positives = 18/27 (66%), Gaps = 3/27 (11%)
Frame = +2
Query: 568 EEDEGEEADGDGDADKDDN---GDDDD 591
E DE +E DG D+DKDD GD+DD
Sbjct: 419 ESDEKKEIDGHEDSDKDDEDEGGDEDD 499
Score = 28.1 bits (61), Expect = 9.9
Identities = 18/70 (25%), Positives = 35/70 (49%), Gaps = 7/70 (10%)
Frame = +1
Query: 51 AEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDED------GGEG 104
A + K+ E +K K + +K + ++ + E+ +++ +EDED G +
Sbjct: 1084 ATSSKTPGETPAKKKKQTSESGKKRKPSDVEEDDQAELSDAKDESQEDEDVAVANNGSDN 1263
Query: 105 E-GEGEGEEE 113
E G+ EGEE+
Sbjct: 1264 EVGKSEGEED 1293
>TC86635 weakly similar to GP|9758600|dbj|BAB09233.1
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1963
Score = 330 bits (846), Expect = 9e-91
Identities = 210/550 (38%), Positives = 319/550 (57%), Gaps = 27/550 (4%)
Frame = +2
Query: 64 KDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEE----- 118
+++G D EEK EVE E +E D K EEE +E++E + + E E EEEEE +E
Sbjct: 5 QEEGND--EEKTEVE-EKEEGDDNEKSEEETEEKEEGDEKEKTEAETEEEEEADEDTIDK 175
Query: 119 ---EEEAEAEADEKVKAKRGPKKGGKKETASEKKD-----PATPASERPTRERKTVERYS 170
E++AE EK KR K +++ +KK+ P TP S+RP RERK+VER
Sbjct: 176 SKEEDKAEGSKGEKGSKKRARGKVNEEKVKVKKKELKLPEPKTPTSDRPVRERKSVERLV 355
Query: 171 VPSPSKSAKSSGGKGLVIEKGRGEQLKDIPNVAFKLSKRKPDDNLHALHSILFGKRTKAY 230
S K L I KGRG LKDIPNVAFKLS+RK DD+L H+ILFG+R KA
Sbjct: 356 ALIDKDSTKE-----LQIAKGRGTPLKDIPNVAFKLSRRKTDDSLKLFHTILFGRRGKAV 520
Query: 231 NLKRNIGQFSGFVWTENEEKQRAKVKERIDKYVKEKLVDFCDVLNIPVSKGNIKKEELSV 290
+K NI +FSGFVW +NEEKQ KVKE++D+ KEKL++FCDVL+I ++K + KE++
Sbjct: 521 QVKSNILRFSGFVWHDNEEKQMLKVKEKLDRCNKEKLLEFCDVLDITITK-STTKEDIIA 697
Query: 291 KLLEFLENPQATTDVLLADNE-KKVKKRTRKVTPSKSPGEASTETPAKKQKKTSE-SGKK 348
KL +FL P ATT V+LAD E K ++RT K +S G +++ K+QKK + SG +
Sbjct: 698 KLTDFLVAPHATTTVVLADKESSKKRRRTVKRGTPRSGGASTSRGSGKRQKKNEDSSGVE 877
Query: 349 RKGSSDVEEDEKAELSDAKDESQENVAAPHKESDDDESKSEEEE-----DKSKAQKKTSK 403
+K ++D E++ + E + +++ + P K D+ KSE E+ +S+ KK +
Sbjct: 878 KKSTTDTEDESEGEEKNEENDDEPENDIPEKSEDETPQKSEREDKSDSGSESEDVKKRKR 1057
Query: 404 KIVKEGSGSKAGERTKSAKKTPVKSSKNVDKTPKKSTPKQTAAEHDSASASLSKSKQPAS 463
+ ++ R+K+ K T S++ P K PK+++ + +++S + S
Sbjct: 1058PSKTSSAKKESAGRSKTEKSTVTNKSRS---PPPKRAPKKSSNTRPKSDDDINESPKVFS 1228
Query: 464 KKRKTE-------NEKPDTKGKASSKKQTDKSSKALVKDQGKSKSGKKAKAVPSREAMHA 516
+K+K+E + TK + SK++T+K +K GK K K+ + P+ + +H
Sbjct: 1229RKKKSEQGGKQKISTPTPTKSSSKSKEKTEKVTK------GKGKK-KETSSSPTDDQLHD 1387
Query: 517 VVVDILKEVDFNTATLSDILRQLGTHFGVDLMHRKAEVKDIITDVINNMSDEEDEGEEAD 576
+ +ILKEVDFNTAT +DIL+ L F VDL +K+ +K +I + +++E DE EE D
Sbjct: 1388AICEILKEVDFNTATFTDILKLLAKQFDVDLTPKKSAIKLMIQKELARLAEEADESEE-D 1564
Query: 577 GDGDADKDDN 586
+ D +KD++
Sbjct: 1565EEEDNEKDED 1594
>CA990541 weakly similar to GP|9758600|dbj
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (13%)
Length = 712
Score = 134 bits (337), Expect = 1e-31
Identities = 97/277 (35%), Positives = 139/277 (50%), Gaps = 8/277 (2%)
Frame = +2
Query: 222 LFGKRTKAYNLKRNIGQFSGFVWTENEEKQRAKVKERIDKYVKEKLVDFCDVLNIPVSKG 281
LF + KA +K+NI +FSGFVW ENEEKQ KVKE+ DK KEKL++F D+L++PV
Sbjct: 2 LFCRIGKAAVIKKNILRFSGFVWHENEEKQMIKVKEKFDKCNKEKLLEFSDLLDVPVGNA 181
Query: 282 NIKKEELSVKLLEFLENPQATTDVLLADNEKKVKKRTRKVTPSKSPGEASTETPAKKQKK 341
N +KE++ KL++FL PQ T VLLA+ EK K++ S G ++ AK +KK
Sbjct: 182 NTRKEDIIAKLIDFLVAPQVTKSVLLAEQEKSAKEKRIAKQGSTGSGTPTSRRSAKSRKK 361
Query: 342 TSES--GKKRKGSSDVEEDEKAELSDAKDESQENVAAPHKESDDDESKSEEEE----DKS 395
+S +K+K ++D E ES+ DE EE E DKS
Sbjct: 362 IEDSSIAEKKKTATDTE----------------------SESEKDEENKEENEIDVSDKS 475
Query: 396 KAQK--KTSKKIVKEGSGSKAGERTKSAKKTPVKSSKNVDKTPKKSTPKQTAAEHDSASA 453
+ +K K K I K+GS S++G T S++++ KN D K T E +S
Sbjct: 476 EDEKPIKGKKHIAKQGS-SRSG--TASSRRSAKSQKKNEDLFVAKKRKTTTDIESES--- 637
Query: 454 SLSKSKQPASKKRKTENEKPDTKGKASSKKQTDKSSK 490
+ E+ K + + S K + KS K
Sbjct: 638 -------------EKEDHKEENENHVSEKSEE*KSIK 709
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 93.2 bits (230), Expect = 3e-19
Identities = 101/493 (20%), Positives = 213/493 (42%), Gaps = 9/493 (1%)
Frame = +1
Query: 29 EVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEV 88
E K E EE + + E+ KS EN E K+ G + EEK +E E E KDE
Sbjct: 1 EEKSHLENEESKETGESSEE------KSHLENEESKETG-ESSEEKSHLENE--ENKDEE 153
Query: 89 KGEEEKDE-EDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGPKKGGKKETASE 147
K ++E +E +D + + E E +EE+ Q+E EE + E + + + +GG+KET
Sbjct: 154 KSKQENEEIKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKETGEI 333
Query: 148 KKDPATPASERPTR-ERKTVERYSVPSPSKSAKSSGGKGLVIEKGRGEQLKDIPNVAFKL 206
++ + +E + K E AK G+ + +G + + K
Sbjct: 334 TEEKSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQGTEETNGTEGGEKE 513
Query: 207 SKRKPDDNLHALHSILFGKRTKAYNLKRNIGQFSGFVWTENEEKQRA-KVKERIDKYVKE 265
K ++ + + + G++ + G + +N+ ++ + K +E+ DK KE
Sbjct: 514 EHDKIKEDTSSDNQVQDGEKNNEAREENYSGDNASSAVVDNKSQESSNKTEEQFDK--KE 687
Query: 266 KLVDFCDVLNIPVSKGNIKKEELSVKLLEFLENPQATTDVLLADNEKKVKKRTRKVTPSK 325
K ++ ++ ++ S + E ++ T + +K + +T TP
Sbjct: 688 K------------NEFELESQKNSNETTESTDS--TITQNSQGNESEKDQAQTENDTPKG 825
Query: 326 SPGEASTETPAKKQKKTSESGKKRKGSSDVE----EDEKAELSDAKDESQENVAAPHKES 381
S E+ + ++Q T++ + +S +++ E S+ + +E+ +A +
Sbjct: 826 SASESDEQKQEQEQNNTTKDDVQTTDTSSQNGNDTTEKQNETSEDANSKKEDSSALNTTP 1005
Query: 382 DDDESKSEEEEDKSKAQKKTSKKIVKEGSGSKAGERTKSAKKTPVKSSKNVDKTPKKSTP 441
++++SKS D++ + TS ++G+ + GE + + P K+S + T + +
Sbjct: 1006NNEDSKSGVAGDQADSTTTTSSSETQDGN-TNHGEYKDTTNENPEKNS-GQEGTQESGSS 1179
Query: 442 KQTAAEHDSAS--ASLSKSKQPASKKRKTENEKPDTKGKASSKKQTDKSSKALVKDQGKS 499
T D+AS L+ + +S+++K E+ ++K ++S + D+ +
Sbjct: 1180SNTFDNKDAASNKVQLTTTSDTSSEQKKDESSSAESKSESSQNDNANSGQSNTTSDESAN 1359
Query: 500 KSGKKAKAVPSRE 512
+ ++ S E
Sbjct: 1360DNKDSSQVTTSSE 1398
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 74.3 bits (181), Expect = 1e-13
Identities = 68/278 (24%), Positives = 115/278 (40%), Gaps = 41/278 (14%)
Frame = +3
Query: 277 PVSKGNIKKEELSVKLLEFLENPQATTDVLLADN----EKKVKKRTRKVTPSKSPGEAST 332
P KGN+KK + P+ T++ +D+ E++VKK K PSK+ +
Sbjct: 366 PAKKGNVKKAQ-----------PETTSEESDSDSSSSDEEEVKKPVSKAVPSKNGSAPAK 512
Query: 333 ETPAKKQKKTSESGKK------------------RKGSSDVEEDEKAELSDAKDESQENV 374
+ +++ + ES + +K +SD E+ +++ D +DE
Sbjct: 513 KVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEEDEKPAAK 692
Query: 375 AAPHK-----------ESDDDESKSEEEEDK---SKAQKKTSKKIVKEGSGSKAGERTKS 420
A P K ES D++S+S +EEDK +KA K S K S S +S
Sbjct: 693 AVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAPTKKAASSSDEESDEES 872
Query: 421 AKKTPVKSSKNVDKTPKKSTPKQTAAEHDSASASLSKSKQP--ASKKRKTENEKPDTKGK 478
+ K KKS + ++ + +S + K+P A K+ K +K +
Sbjct: 873 DEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVAAKKEDKMNVDKDGSDSD 1052
Query: 479 ASSKKQTDKSSKA---LVKDQGKSKSGKKAKAVPSREA 513
S ++ D+ SK +KD +GK K P+ A
Sbjct: 1053QSEEESEDEPSKTPQKKIKDVEMVDAGKSGKKAPNTPA 1166
Score = 68.9 bits (167), Expect = 5e-12
Identities = 99/426 (23%), Positives = 163/426 (38%), Gaps = 25/426 (5%)
Frame = +3
Query: 55 KSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEE 114
KS ++A + D V + +K + ++EVK K ++ E+ + + ++E
Sbjct: 108 KSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQKVEEVAAKQKALKVVKKE 287
Query: 115 EQEEEEEAEAEADE----------KVKAKRGPKKGGKKETASEKKDPATPASERPTRERK 164
E EE +E+E ++ K AK+G K + ET SE+ D + +S+ E +
Sbjct: 288 ESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSD----EEE 455
Query: 165 TVERYSVPSPSKSAKSSGGKGLVIEKGRGEQLKDIPNVAFKLSKRKPDDNLHALHSILFG 224
+ S PSK+ + K E+ E+ D +K P N A
Sbjct: 456 VKKPVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKP--AAKAVPSKNGSA------- 608
Query: 225 KRTKAYNLKRNIGQFSGFVWTENEEKQRAKVKERIDKYVKEKLVDFCDVLNIPVSKGNIK 284
KA + + + + S E+EE ++ K +P G++
Sbjct: 609 PAKKAASDEEDTDESSD----EDEEDEKPAAKA------------------VPSKNGSV- 719
Query: 285 KEELSVKLLEFLENPQATTDVLLADNEKKVKKRTRKVTPSKSPGEASTETPAKKQKKTSE 344
P D +D + S+S E + AK K S
Sbjct: 720 --------------PAKKADTESSDED------------SESSDEEDKKPAAKASKNVSA 821
Query: 345 SGKKRKGSSDVEEDEKA-ELSDAKDESQENVAAPHKESD-------DDESKSEEEEDKSK 396
KK SSD E DE++ E DAK S+ A + D DD+S S+E++
Sbjct: 822 PTKKAASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVA 1001
Query: 397 AQKKTSKKIVKEGSGSKAGERTK--SAKKTPVKSSKNVD-----KTPKKSTPKQTAAEHD 449
A+K+ + K+GS S E KTP K K+V+ K+ KK+ +
Sbjct: 1002AKKEDKMNVDKDGSDSDQSEEESEDEPSKTPQKKIKDVEMVDAGKSGKKAPNTPATPIEN 1181
Query: 450 SASASL 455
S S +L
Sbjct: 1182SGSKTL 1199
Score = 57.0 bits (136), Expect = 2e-08
Identities = 77/291 (26%), Positives = 119/291 (40%), Gaps = 19/291 (6%)
Frame = +3
Query: 321 VTPSKSPGEASTETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHKE 380
+ P S A+ A +SGKK G EE+ KA AK + E VAA K
Sbjct: 99 IMPKSSKKSATKVDAAPVVVSPVKSGKK--GKRQAEEEVKAV--SAKKQKVEEVAAKQKA 266
Query: 381 SDDDESKSEEEEDKSKAQKKTSKKIVKEGSGSKAGERTKSAKKTPVKSSKNVDKTPKKST 440
K EE + ++ + + +VK + SK KTP K NV KK+
Sbjct: 267 LK--VVKKEESSSEESSESEDEQPVVKAPAPSK---------KTPAKKG-NV----KKAQ 398
Query: 441 PKQTAAEHDSASASLSKS--KQPASKKRKTEN-EKPDTKGKASSKKQTDKSSKALVKDQG 497
P+ T+ E DS S+S + K+P SK ++N P K S ++ +++SS D+
Sbjct: 399 PETTSEESDSDSSSSDEEEVKKPVSKAVPSKNGSAPAKKVDTSEEEDSEESS-----DED 563
Query: 498 KSKSGKKAKAVPSREAMHAVVVDILKEVDFNTATLSDILRQLGTHFGVDLMHRKAEVKDI 557
K + AKAVPS+ E D + ++ D + V + K
Sbjct: 564 KKPA---AKAVPSKNGSAPAKKAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKA 734
Query: 558 IT---DVINNMSDEEDE-------------GEEADGDGDADKDDNGDDDDE 592
T D + SDEED+ ++A D + D+ D+D++
Sbjct: 735 DTESSDEDSESSDEEDKKPAAKASKNVSAPTKKAASSSDEESDEESDEDED 887
Score = 55.8 bits (133), Expect = 4e-08
Identities = 42/169 (24%), Positives = 72/169 (41%), Gaps = 16/169 (9%)
Frame = +3
Query: 14 VQAEGDDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEE 73
V A+ D +++++E E E+K+ A + ++ SA K+ + + EE D+ D+ E+
Sbjct: 717 VPAKKADTESSDEDSESSDE---EDKKPAAKASKNVSAPTKKAASSSDEESDEESDEDED 887
Query: 74 KEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEG----------------EGEGEEEEEQE 117
+ V K K K + D+ED+D E +G ++ E+E
Sbjct: 888 AKPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVAAKKEDKMNVDKDGSDSDQSEEE 1067
Query: 118 EEEEAEAEADEKVKAKRGPKKGGKKETASEKKDPATPASERPTRERKTV 166
E+E +K+K G S KK P TPA+ KT+
Sbjct: 1068SEDEPSKTPQKKIKDVEMVDAG-----KSGKKAPNTPATPIENSGSKTL 1199
Score = 53.5 bits (127), Expect = 2e-07
Identities = 42/176 (23%), Positives = 73/176 (40%), Gaps = 11/176 (6%)
Frame = +3
Query: 17 EGDDAAPNNQETEVK-----------GEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKD 65
E D + ++ E EVK G A A++ + + +ED + S++ K A A
Sbjct: 417 ESDSDSSSSDEEEVKKPVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSK 596
Query: 66 KGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAE 125
G ++ E++TDE DE + +E+ + G + + E +E+ E+ E
Sbjct: 597 NGSAPAKKAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDE 776
Query: 126 ADEKVKAKRGPKKGGKKETASEKKDPATPASERPTRERKTVERYSVPSPSKSAKSS 181
D+K A K K +A KK ++ E + + V P+ AK S
Sbjct: 777 EDKKPAA-----KASKNVSAPTKKAASSSDEESDEESDEDEDAKPVSKPAAVAKKS 929
Score = 45.1 bits (105), Expect = 8e-05
Identities = 40/173 (23%), Positives = 74/173 (42%), Gaps = 10/173 (5%)
Frame = +3
Query: 19 DDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVE 78
++ A + +V + E+ +E++ EDE VK+ A + + K G+ + + E
Sbjct: 240 EEVAAKQKALKVVKKEESSSEESSESEDEQP---VVKAPAPSKKTPAKKGNVKKAQPETT 410
Query: 79 KETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADE--KVKAKRGP 136
E + EEE + + ++ + EEE++E +DE K AK P
Sbjct: 411 SEESDSDSSSSDEEEVKKPVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVP 590
Query: 137 KKGGK---KETASEKKDPATPA-----SERPTRERKTVERYSVPSPSKSAKSS 181
K G K+ AS+++D + E+P + + SVP+ +SS
Sbjct: 591 SKNGSAPAKKAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESS 749
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 68.9 bits (167), Expect = 5e-12
Identities = 52/180 (28%), Positives = 89/180 (48%), Gaps = 15/180 (8%)
Frame = +2
Query: 17 EGDDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEK-- 74
+ DD ++ + KG +E E+ + ++DED + + E+ +EK KGG +E+K
Sbjct: 413 DDDDDFEGVEKKKAKGGSEGED-DFEEEDDEDEEGSEDEDDEEDEKEKVKGGG-IEDKFL 586
Query: 75 ------EEVEKETDEV-KDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEAD 127
E +EKE D K E + E ++D E++D E GE E ++E++ +++EE E EA+
Sbjct: 587 KIDELTEYLEKEEDNYEKGEERDEADEDSEEDDELEKAGEFEMDDEDDDDDDEEDE-EAE 763
Query: 128 EKVKAKRGPKKGGKKETASEKKDPATPAS------ERPTRERKTVERYSVPSPSKSAKSS 181
+ + GGK E S++KD S E ++++T Y S K S
Sbjct: 764 DMGNVRYEDFFGGKNEKGSKRKDQLLEVSGDEDDMESTKQKKRTASTYEKQRGKNSIKDS 943
Score = 50.4 bits (119), Expect = 2e-06
Identities = 28/90 (31%), Positives = 51/90 (56%)
Frame = +2
Query: 46 DEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGE 105
+E A + + E+K + + EE ++ ++E D+ D+ +G E+K + + G EGE
Sbjct: 302 EEIAQFKVPLDVGKKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKK--KAKGGSEGE 475
Query: 106 GEGEGEEEEEQEEEEEAEAEADEKVKAKRG 135
+ E E++E++E E+ + E DEK K K G
Sbjct: 476 DDFEEEDDEDEEGSEDEDDEEDEKEKVKGG 565
Score = 47.8 bits (112), Expect = 1e-05
Identities = 28/78 (35%), Positives = 44/78 (55%), Gaps = 5/78 (6%)
Frame = +2
Query: 68 GDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGE-----GEGEGEGEEEEEQEEEEEA 122
G ++E+K+ VE E +E D+ E + D++D +G E G EGE + EEE +E+EE
Sbjct: 338 GKKLEKKKRVELEEEE-SDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEEDDEDEEG 514
Query: 123 EAEADEKVKAKRGPKKGG 140
+ D++ K K GG
Sbjct: 515 SEDEDDEEDEKEKVKGGG 568
Score = 30.8 bits (68), Expect = 1.5
Identities = 51/225 (22%), Positives = 88/225 (38%), Gaps = 30/225 (13%)
Frame = +2
Query: 255 VKERIDKYVK--EKLVDFCDVLNIPVSKGNIKKEELSVKLLEFLENPQATTDVLLADNEK 312
++ R++++VK E++ F +P+ G KK E K +E E D L D++
Sbjct: 266 LRRRLNQFVKNPEEIAQF----KVPLDVG--KKLEKK-KRVELEEEESDDFDEELDDDDD 424
Query: 313 KVKKRTRKVTPSKSPGEASTETPAKKQKKTSESG-------KKRKGSS------------ 353
+ +K S GE E + ++ SE +K KG
Sbjct: 425 DFEGVEKKKAKGGSEGEDDFEEEDDEDEEGSEDEDDEEDEKEKVKGGGIEDKFLKIDELT 604
Query: 354 ---DVEED--EKAELSDAKDESQENVAAPHK----ESDDDESKSEEEEDKSKAQKKTSKK 404
+ EED EK E D DE E K E DD++ ++EED+ +
Sbjct: 605 EYLEKEEDNYEKGEERDEADEDSEEDDELEKAGEFEMDDEDDDDDDEEDEEAEDMGNVRY 784
Query: 405 IVKEGSGSKAGERTKSAKKTPVKSSKNVDKTPKKSTPKQTAAEHD 449
G ++ G + K +++ T +K K+TA+ ++
Sbjct: 785 EDFFGGKNEKGSKRKDQLLEVSGDEDDMESTKQK---KRTASTYE 910
Score = 28.9 bits (63), Expect = 5.8
Identities = 14/33 (42%), Positives = 23/33 (69%)
Frame = +2
Query: 560 DVINNMSDEEDEGEEADGDGDADKDDNGDDDDE 592
D + S+E+DE E+A G+ + D +D+ DDD+E
Sbjct: 653 DEADEDSEEDDELEKA-GEFEMDDEDDDDDDEE 748
>BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfalfa, partial
(29%)
Length = 663
Score = 66.6 bits (161), Expect = 3e-11
Identities = 61/204 (29%), Positives = 93/204 (44%), Gaps = 13/204 (6%)
Frame = +2
Query: 321 VTPSKSPGEASTETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHK- 379
+ P S A+ A +SGKK G EE+ KA AK + E VAA K
Sbjct: 77 IMPKSSKKSATKVDAAPVVVSPVKSGKK--GKRQAEEEVKAV--SAKKQKVEEVAAKQKA 244
Query: 380 --------ESDDDESKSEEEEDKSKAQKKTSKKIVKEGSGSKAGERTKSAKKTPVKSSKN 431
S ++ S+SE+E+ KA + K K+G+ KA T S + SS +
Sbjct: 245 LKVVKKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSD 424
Query: 432 VDKTPKKSTPKQTAAEHDSASA----SLSKSKQPASKKRKTENEKPDTKGKASSKKQTDK 487
++ KK K +++ SA A + + K+PA+K ++N K AS ++ TD+
Sbjct: 425 EEEV-KKPVSKAVPSKNGSAPAKKVDTSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDE 601
Query: 488 SSKALVKDQGKSKSGKKAKAVPSR 511
SS D+ + AKAVPS+
Sbjct: 602 SS-----DEDEEXXKPAAKAVPSK 658
Score = 43.1 bits (100), Expect = 3e-04
Identities = 40/132 (30%), Positives = 57/132 (42%), Gaps = 4/132 (3%)
Frame = +2
Query: 277 PVSKGNIKKEELSVKLLEFLENPQATTDVLLADN----EKKVKKRTRKVTPSKSPGEAST 332
P KGN+KK + P+ T++ +D+ E++VKK K PSK+ G A
Sbjct: 344 PAKKGNVKKAQ-----------PETTSEESDSDSSSSDEEEVKKPVSKAVPSKN-GSA-- 481
Query: 333 ETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHKESDDDESKSEEEE 392
PAKK +SD ++ A+ +K+ S A E D DES E+EE
Sbjct: 482 --PAKK-----------VDTSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEE 622
Query: 393 DKSKAQKKTSKK 404
A K K
Sbjct: 623 XXKPAAKAVPSK 658
Score = 38.5 bits (88), Expect = 0.007
Identities = 38/157 (24%), Positives = 69/157 (43%), Gaps = 10/157 (6%)
Frame = +2
Query: 55 KSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEG------EG 108
KS ++A + D V + +K + ++EVK K ++ E+ + +
Sbjct: 86 KSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQKVEEVAAKQKALKVVKKE 265
Query: 109 EGEEEEEQEEEEE---AEAEA-DEKVKAKRGPKKGGKKETASEKKDPATPASERPTRERK 164
E EE E E+E +A A +K AK+G K + ET SE+ D + +S+ E +
Sbjct: 266 ESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSD----EEE 433
Query: 165 TVERYSVPSPSKSAKSSGGKGLVIEKGRGEQLKDIPN 201
+ S PSK+ + K ++ + K +P+
Sbjct: 434 VKKPVSKAVPSKNGSAPAKKVDTSDEDKKPAAKAVPS 544
Score = 34.7 bits (78), Expect = 0.11
Identities = 42/200 (21%), Positives = 69/200 (34%), Gaps = 15/200 (7%)
Frame = +2
Query: 399 KKTSKKIVKEGSGSKAGERTKSAKKTPVKSSKNVDKTPKKSTPKQTAAEHDSASASLSKS 458
K + K K + KS KK ++ + V K + A A + K
Sbjct: 86 KSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQKVEEVAAKQKALKVVKKE 265
Query: 459 KQPASKKRKTENEKPDTKGKASSKK------QTDKSSKALVKDQGKSKSGKK-------- 504
+ + + ++E+E+P K A SKK K+ ++ S S
Sbjct: 266 ESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEVKKP 445
Query: 505 -AKAVPSREAMHAVVVDILKEVDFNTATLSDILRQLGTHFGVDLMHRKAEVKDIITDVIN 563
+KAVPS+ K+VD + + + + G + A
Sbjct: 446 VSKAVPSKNGSAPA-----KKVDTSDEDKKPAAKAVPSKNGSAPAKKAA----------- 577
Query: 564 NMSDEEDEGEEADGDGDADK 583
SDEED E +D D + K
Sbjct: 578 --SDEEDTDESSDEDEEXXK 631
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 66.2 bits (160), Expect = 3e-11
Identities = 71/290 (24%), Positives = 121/290 (41%), Gaps = 21/290 (7%)
Frame = +3
Query: 15 QAEGDDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEK--DKGGD-QV 71
Q EG++ N ETE + E E+ E+E+ V+ E E+ E+ D+GGD ++
Sbjct: 468 QQEGEEG--NKHETEEESEDNVHERREEQDEEENKHGAEVQEENESKSEEVEDEGGDVEI 641
Query: 72 EEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEE-------EQEEEEEAEA 124
+E + + E D +++ +EEKD+E+E E E E + +EE+ E E E
Sbjct: 642 DENDHEKSEADNDREDEVVDEEKDKEEEGDDETENEDKEDEEKGGLVENHENHEAREEHY 821
Query: 125 EADEKVKAKRGPKKGGKKETAS-EKKDPATPASERPTRERKTVERYSVPSPSKSAKSSGG 183
+AD+ A ET + E D + + +P E + K + G
Sbjct: 822 KADDASSAVAHDTHETSTETGNLEHSDLSLQNTRKPENETNHSDESYGSQNVSDLKVTEG 1001
Query: 184 KGLVIEKGRGEQLKDIPNVAF--KLSKRKPDDNLHALHSILFGKRTKAYNLKRNIG---- 237
+ K+ N +F + +K KPD L L S L T+A + G
Sbjct: 1002ELTDGVSSNATAGKETGNDSFSNETAKTKPDSQLD-LSSNLTAVITEASSNSSGTGDDTS 1178
Query: 238 ----QFSGFVWTENEEKQRAKVKERIDKYVKEKLVDFCDVLNIPVSKGNI 283
Q + +E++ Q A V I +K+ + + +S+GN+
Sbjct: 1179SSSEQIKAVILSESDHAQNATVNTTITGDMKQ--TEGLEQSGSKISEGNL 1322
Score = 54.7 bits (130), Expect = 1e-07
Identities = 48/160 (30%), Positives = 75/160 (46%), Gaps = 9/160 (5%)
Frame = +3
Query: 15 QAEGDDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENA-EEKDKGGDQVEE 73
+ E + N Q E + E +E E E+ S + V E EE++K G +V+E
Sbjct: 408 EEEDEHIVYNMQNKREHDEQQQEGEEGNKHETEEESEDNVHERREEQDEEENKHGAEVQE 587
Query: 74 K-----EEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADE 128
+ EEVE E +V+ + + + EK E D D + + E ++EEE ++E E E + DE
Sbjct: 588 ENESKSEEVEDEGGDVEID-ENDHEKSEADNDREDEVVDEEKDKEEEGDDETENEDKEDE 764
Query: 129 KVKAKRGPKKGGKKETASE---KKDPATPASERPTRERKT 165
+ K G + + A E K D A+ A T E T
Sbjct: 765 E---KGGLVENHENHEAREEHYKADDASSAVAHDTHETST 875
Score = 40.0 bits (92), Expect = 0.003
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 1/86 (1%)
Frame = +3
Query: 64 KDKGGDQVE-EKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEA 122
KD +VE +K E +E +E + V + K E DE EGE + E EEE E+
Sbjct: 354 KDLHPGKVEADKNEGHEEEEEDEHIVYNMQNKREHDEQQQEGEEGNKHETEEESEDNVHE 533
Query: 123 EAEADEKVKAKRGPKKGGKKETASEK 148
E ++ + K G + + E+ SE+
Sbjct: 534 RREEQDEEENKHGAEVQEENESKSEE 611
Score = 40.0 bits (92), Expect = 0.003
Identities = 68/324 (20%), Positives = 118/324 (35%), Gaps = 35/324 (10%)
Frame = +3
Query: 224 GKRTKAYNLKRNIGQFSGFVWTENEEKQRAKVKERIDKYVKEKLVDFCDVLNIPVSKGNI 283
G TK +KRN + N+ + KVK + + L+ C L V +
Sbjct: 126 GFLTKLSMIKRNPS-------SRNQRSKGFKVKHVLQAIL---LLGVCFWLIYQVKHNHD 275
Query: 284 KKEELSVKLLEFLENPQATTDVLLADNEKKVKKRTRKVTPSKSPGEASTETPAKKQKKTS 343
KK+E + + TD +L K + KV K+ G E + +
Sbjct: 276 KKKEFDKNDTKLPIRTE--TDQILKLGRKDL--HPGKVEADKNEGHEEEE---EDEHIVY 434
Query: 344 ESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHKESDDDE------------SKSEEE 391
KR+ +E E+ + ++ES++NV +E D++E SKSEE
Sbjct: 435 NMQNKREHDEQQQEGEEGNKHETEEESEDNVHERREEQDEEENKHGAEVQEENESKSEEV 614
Query: 392 ED-------------KSKAQKKTSKKIV-------KEGSGSKAGERTKSAKKTP-VKSSK 430
ED KS+A ++V +EG E + +K V++ +
Sbjct: 615 EDEGGDVEIDENDHEKSEADNDREDEVVDEEKDKEEEGDDETENEDKEDEEKGGLVENHE 794
Query: 431 NVDKTPK--KSTPKQTAAEHDSASASLSKSKQPASKKRKTENEKPDTKGKASSKKQTDKS 488
N + + K+ +A HD+ S S KP+ + S + ++
Sbjct: 795 NHEAREEHYKADDASSAVAHDTHETSTETGNLEHSDLSLQNTRKPENETNHSDESYGSQN 974
Query: 489 SKALVKDQGKSKSGKKAKAVPSRE 512
L +G+ G + A +E
Sbjct: 975 VSDLKVTEGELTDGVSSNATAGKE 1046
Score = 33.1 bits (74), Expect = 0.31
Identities = 36/186 (19%), Positives = 67/186 (35%), Gaps = 44/186 (23%)
Frame = +3
Query: 57 EAENAEEKDKGGDQVEEKEEVEKETDEV--------------KDEVKGEEEKDEEDE--- 99
+ ++ +K K D+ + K + ETD++ D+ +G EE++E++
Sbjct: 255 QVKHNHDKKKEFDKNDTKLPIRTETDQILKLGRKDLHPGKVEADKNEGHEEEEEDEHIVY 434
Query: 100 ------DGGEGEGEGEGEEEEEQEEEEE------------------AEAEADEKVKAKRG 135
+ E + EGE + E EEE E AE + + + K++
Sbjct: 435 NMQNKREHDEQQQEGEEGNKHETEEESEDNVHERREEQDEEENKHGAEVQEENESKSEEV 614
Query: 136 PKKGGK---KETASEKKDPATPASERPTRERKTVERYSVPSPSKSAKSSGGKGLVIEKGR 192
+GG E EK + + E K E K KG ++E
Sbjct: 615 EDEGGDVEIDENDHEKSEADNDREDEVVDEEKDKEEEGDDETENEDKEDEEKGGLVENHE 794
Query: 193 GEQLKD 198
+ ++
Sbjct: 795 NHEARE 812
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 65.1 bits (157), Expect = 7e-11
Identities = 33/78 (42%), Positives = 50/78 (63%)
Frame = +3
Query: 59 ENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEE 118
E+ EE+D+G + EE E E+E DE +E + EEE++EEDE E E E E EEEEE+++
Sbjct: 48 EHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDD 227
Query: 119 EEEAEAEADEKVKAKRGP 136
+EE E + +++ P
Sbjct: 228 DEEGEEDEIDRISTLPDP 281
Score = 58.2 bits (139), Expect = 9e-09
Identities = 30/71 (42%), Positives = 48/71 (67%)
Frame = +3
Query: 58 AENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQE 117
AE++ ++ K + EE++E E E +E + + + E+E DEE+E+ E E E E ++EEE+E
Sbjct: 15 AESSWKRRKVEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEE 194
Query: 118 EEEEAEAEADE 128
EEEE E E D+
Sbjct: 195 EEEEEEEEEDD 227
Score = 55.5 bits (132), Expect = 6e-08
Identities = 27/75 (36%), Positives = 46/75 (61%)
Frame = +3
Query: 33 EAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEE 92
E+ + ++ +E+ E + E E+ EE++ D+ EE+EE E+E DE DE + EE
Sbjct: 18 ESSWKRRKVEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEE 197
Query: 93 EKDEEDEDGGEGEGE 107
E++EE+E+ + EGE
Sbjct: 198 EEEEEEEEDDDEEGE 242
Score = 50.8 bits (120), Expect = 1e-06
Identities = 30/74 (40%), Positives = 46/74 (61%)
Frame = +3
Query: 70 QVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEK 129
+VEE E E E EV++E E+E DEE+ED + E E E EEEEE E ++E E E +E+
Sbjct: 39 KVEEHNEEEDE-GEVEEE---EDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEE 206
Query: 130 VKAKRGPKKGGKKE 143
+ + + G+++
Sbjct: 207 EEEEEDDDEEGEED 248
Score = 47.8 bits (112), Expect = 1e-05
Identities = 28/77 (36%), Positives = 46/77 (59%)
Frame = +3
Query: 74 KEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAK 133
K +E +E +DE + EEE+DE DE E E E +EEEE+EEEEE E E D++ + +
Sbjct: 30 KRRKVEEHNEEEDEGEVEEEEDEHDE-----EEEDEHDEEEEEEEEEEEEDEHDDEEEEE 194
Query: 134 RGPKKGGKKETASEKKD 150
++ +++ E ++
Sbjct: 195 EEEEEEEEEDDDEEGEE 245
Score = 42.4 bits (98), Expect = 5e-04
Identities = 28/76 (36%), Positives = 39/76 (50%), Gaps = 1/76 (1%)
Frame = +3
Query: 25 NQETEVKGEAEAEEKEA-APQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDE 83
+ E E +GE E EE E +EDE E + E E +E D ++ EE+EE E+E D
Sbjct: 51 HNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDD- 227
Query: 84 VKDEVKGEEEKDEEDE 99
+E+ EEDE
Sbjct: 228 --------DEEGEEDE 251
Score = 40.0 bits (92), Expect = 0.003
Identities = 26/87 (29%), Positives = 42/87 (47%), Gaps = 1/87 (1%)
Frame = +3
Query: 89 KGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGPKKGGKKETASEK 148
K EE +EEDE GE E EE+E EEEE+ E +E+ + + + +E E+
Sbjct: 39 KVEEHNEEEDE------GEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEE 200
Query: 149 KDPATPASERPTRERKTVERYS-VPSP 174
++ + E ++R S +P P
Sbjct: 201 EEEEEEEDDDEEGEEDEIDRISTLPDP 281
Score = 36.6 bits (83), Expect = 0.028
Identities = 16/68 (23%), Positives = 38/68 (55%)
Frame = +3
Query: 333 ETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHKESDDDESKSEEEE 392
E+ K++K + ++ +G + EEDE E + + + +E +E D+ + + EEEE
Sbjct: 18 ESSWKRRKVEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEE 197
Query: 393 DKSKAQKK 400
++ + +++
Sbjct: 198 EEEEEEEE 221
Score = 34.3 bits (77), Expect = 0.14
Identities = 18/74 (24%), Positives = 37/74 (49%)
Frame = +3
Query: 320 KVTPSKSPGEASTETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHK 379
K+T ++S + +++ E ++ + EEDE E + ++E +E +
Sbjct: 3 KMTMAESSWKRRKVEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDE 182
Query: 380 ESDDDESKSEEEED 393
E +++E + EEEED
Sbjct: 183 EEEEEEEEEEEEED 224
>TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (15%)
Length = 557
Score = 65.1 bits (157), Expect = 7e-11
Identities = 40/110 (36%), Positives = 63/110 (56%), Gaps = 3/110 (2%)
Frame = +2
Query: 37 EEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDE 96
E KE ++E EA K + EEK++G D EEK EVE E +E D K EEE +E
Sbjct: 83 EVKETTEDKEEKEKIEAEKEAEGSIEEKEEGND--EEKTEVE-EKEEGDDNEKSEEETEE 253
Query: 97 EDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGPKK---GGKKE 143
++E + + E E EEEEE +E+ +++ ++K + +G K+ GG+ +
Sbjct: 254 KEEGDEKEKTEAETEEEEEADEDTIDKSKEEDKAEGSKGGKE***GGRSK 403
Score = 47.8 bits (112), Expect = 1e-05
Identities = 29/90 (32%), Positives = 50/90 (55%), Gaps = 3/90 (3%)
Frame = +2
Query: 26 QETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKD---KGGDQVEEKEEVEKETD 82
++ E K + EAE++ E+++ + K+E E EE D K ++ EEKEE + E +
Sbjct: 101 EDKEEKEKIEAEKEAEGSIEEKEEGNDEEKTEVEEKEEGDDNEKSEEETEEKEEGD-EKE 277
Query: 83 EVKDEVKGEEEKDEEDEDGGEGEGEGEGEE 112
+ + E + EEE DE+ D + E + EG +
Sbjct: 278 KTEAETEEEEEADEDTIDKSKEEDKAEGSK 367
Score = 46.2 bits (108), Expect = 4e-05
Identities = 28/103 (27%), Positives = 51/103 (49%)
Frame = +2
Query: 83 EVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGPKKGGKK 142
EVK+ + +EEK++ E E E EG EE++E +E + E +EK + K ++
Sbjct: 83 EVKETTEDKEEKEKI-----EAEKEAEGSIEEKEEGNDEEKTEVEEKEEGDDNEK--SEE 241
Query: 143 ETASEKKDPATPASERPTRERKTVERYSVPSPSKSAKSSGGKG 185
ET +++ +E T E + + ++ + K+ G KG
Sbjct: 242 ETEEKEEGDEKEKTEAETEEEEEADEDTIDKSKEEDKAEGSKG 370
Score = 45.1 bits (105), Expect = 8e-05
Identities = 30/97 (30%), Positives = 55/97 (55%)
Frame = +2
Query: 54 VKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEE 113
VK E+ EEK+K +E ++E E +E K+E EE+ + E+++ EG+ + E
Sbjct: 86 VKETTEDKEEKEK----IEAEKEAEGSIEE-KEEGNDEEKTEVEEKE----EGDDNEKSE 238
Query: 114 EEQEEEEEAEAEADEKVKAKRGPKKGGKKETASEKKD 150
EE EE+EE + + EK +A+ ++ ++T + K+
Sbjct: 239 EETEEKEEGDEK--EKTEAETEEEEEADEDTIDKSKE 343
Score = 44.3 bits (103), Expect = 1e-04
Identities = 24/86 (27%), Positives = 46/86 (52%)
Frame = +2
Query: 27 ETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKD 86
E E + E EEKE DE+ + K E ++ E+ ++ ++ EE +E EK E ++
Sbjct: 128 EAEKEAEGSIEEKEEG--NDEEKTEVEEKEEGDDNEKSEEETEEKEEGDEKEKTEAETEE 301
Query: 87 EVKGEEEKDEEDEDGGEGEGEGEGEE 112
E + +E+ ++ ++ + EG G+E
Sbjct: 302 EEEADEDTIDKSKEEDKAEGSKGGKE 379
Score = 42.4 bits (98), Expect = 5e-04
Identities = 24/66 (36%), Positives = 39/66 (58%), Gaps = 2/66 (3%)
Frame = +2
Query: 7 EDHNKPPVQAEGDDAAPNNQETEVKGEAEAEEK-EAAPQEDEDASAEAV-KSEAENAEEK 64
E+ + + EGDD + +ETE K E + +EK EA +E+E+A + + KS+ E+ E
Sbjct: 182 EEKTEVEEKEEGDDNEKSEEETEEKEEGDEKEKTEAETEEEEEADEDTIDKSKEEDKAEG 361
Query: 65 DKGGDQ 70
KGG +
Sbjct: 362 SKGGKE 379
Score = 37.4 bits (85), Expect = 0.016
Identities = 23/95 (24%), Positives = 46/95 (48%), Gaps = 4/95 (4%)
Frame = +2
Query: 309 DNEKKVKKRTRKVTPS----KSPGEASTETPAKKQKKTSESGKKRKGSSDVEEDEKAELS 364
D E+K K K K G +T +++++ ++ K + + + EE ++ E +
Sbjct: 104 DKEEKEKIEAEKEAEGSIEEKEEGNDEEKTEVEEKEEGDDNEKSEEETEEKEEGDEKEKT 283
Query: 365 DAKDESQENVAAPHKESDDDESKSEEEEDKSKAQK 399
+A+ E +E E+D+D +EEDK++ K
Sbjct: 284 EAETEEEE-------EADEDTIDKSKEEDKAEGSK 367
Score = 37.0 bits (84), Expect = 0.021
Identities = 28/101 (27%), Positives = 48/101 (46%), Gaps = 13/101 (12%)
Frame = +2
Query: 329 EASTETPAKKQKKTSESGKKRKGSSDVEED----------EKAELSD---AKDESQENVA 375
E T K++K+ E+ K+ +GS + +E+ EK E D +++E++E
Sbjct: 83 EVKETTEDKEEKEKIEAEKEAEGSIEEKEEGNDEEKTEVEEKEEGDDNEKSEEETEEKEE 262
Query: 376 APHKESDDDESKSEEEEDKSKAQKKTSKKIVKEGSGSKAGE 416
KE + E++ EEE D+ K K + GSK G+
Sbjct: 263 GDEKEKTEAETEEEEEADEDTIDK---SKEEDKAEGSKGGK 376
Score = 31.6 bits (70), Expect = 0.89
Identities = 19/70 (27%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Frame = +3
Query: 63 EKDKGGDQVEEKEEVEKETDEVKDEVKGEEEK----DEEDEDGGEGEGEGEGEEEEEQEE 118
+K K +V + E+ + E +EVKD+++ + K D+ D+D + E + E + E
Sbjct: 336 QKKKIRPRVVKVEKNDNEVEEVKDDMEVDSVKEPVEDKNDDDDIQEMEEDDKNVAESENE 515
Query: 119 EEEAEAEADE 128
+ + +AE E
Sbjct: 516 KMDVDAEVKE 545
Score = 29.6 bits (65), Expect = 3.4
Identities = 27/94 (28%), Positives = 48/94 (50%), Gaps = 2/94 (2%)
Frame = +3
Query: 312 KKVKKRTRKVTPSKSPGEASTETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQ 371
K+ KR RK+ + T QKK K R VE+++ E+ + KD+ +
Sbjct: 258 KRAMKR-RKLKQRRRRRRRPTRILLINQKK-----KIRPRVVKVEKNDN-EVEEVKDDME 416
Query: 372 -ENVAAPHKE-SDDDESKSEEEEDKSKAQKKTSK 403
++V P ++ +DDD+ + EE+DK+ A+ + K
Sbjct: 417 VDSVKEPVEDKNDDDDIQEMEEDDKNVAESENEK 518
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 65.1 bits (157), Expect = 7e-11
Identities = 43/156 (27%), Positives = 86/156 (54%), Gaps = 7/156 (4%)
Frame = +3
Query: 17 EGDDAAPNNQETEVKGEAEAEE---KEAAPQEDEDASAEAVKSEAENA-EEKDKGGDQVE 72
E DD ++ E K + + +E K+ ++ ++ E VK E E+ E+KDK + E
Sbjct: 168 EKDDEKKEKKDKEKKDKTDVDEGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKE 347
Query: 73 EKEEVEKETD---EVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEK 129
+KE+ +++ D E K K +E+K +++ED EGE +G ++++ ++++E + E DEK
Sbjct: 348 KKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDEGE---DGSKKKKNKDKKEKKKEEDEK 518
Query: 130 VKAKRGPKKGGKKETASEKKDPATPASERPTRERKT 165
+ K + +ETA E K+ ++ +++K+
Sbjct: 519 EEGKVSVRDIDIEETAKEGKEKKKKKEDKEEKKKKS 626
Score = 63.5 bits (153), Expect = 2e-10
Identities = 43/150 (28%), Positives = 78/150 (51%), Gaps = 4/150 (2%)
Frame = +3
Query: 4 ETLEDHNKPPVQAEGDDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEE 63
E E +K D + ++ E K + + EE +ED D E E + E+
Sbjct: 180 EKKEKKDKEKKDKTDVDEGKDKKDKEKKKKEKKEENVKGEEEDGD---EKKDKEKKKKEK 350
Query: 64 KDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEE-- 121
K+KG + ++K+ EK++ + K++ K + E D+E EDG + + + +E++++E+E+E
Sbjct: 351 KEKGKED-KDKDGEEKKSKKDKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEG 527
Query: 122 --AEAEADEKVKAKRGPKKGGKKETASEKK 149
+ + D + AK G +K KKE EKK
Sbjct: 528 KVSVRDIDIEETAKEGKEKKKKKEDKEEKK 617
Score = 61.6 bits (148), Expect = 8e-10
Identities = 35/131 (26%), Positives = 72/131 (54%)
Frame = +3
Query: 20 DAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEK 79
D + E E E ++ EK+ +E +D + E ++KDK + E+KEE K
Sbjct: 114 DVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKKDKEKKKKEKKEENVK 293
Query: 80 ETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGPKKG 139
+E DE K +E+K +E ++ G+ + + +GEE++ ++++E+ + + ++ + + G KK
Sbjct: 294 GEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDEGEDGSKKK 473
Query: 140 GKKETASEKKD 150
K+ +KK+
Sbjct: 474 KNKDKKEKKKE 506
Score = 60.1 bits (144), Expect = 2e-09
Identities = 40/151 (26%), Positives = 80/151 (52%), Gaps = 7/151 (4%)
Frame = +3
Query: 24 NNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGG------DQVEEKEEV 77
+N ++ ++ + KE + ++D ++V+ + E E+KDK D+ ++K++
Sbjct: 75 SNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKKDK 254
Query: 78 EKETDEVKDE-VKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGP 136
EK+ E K+E VKGEE EDG E + + + ++E++++ +E+ + + +EK K
Sbjct: 255 EKKKKEKKEENVKGEE------EDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKE 416
Query: 137 KKGGKKETASEKKDPATPASERPTRERKTVE 167
KK K E E +D + + +E+K E
Sbjct: 417 KKKDKNEDDDEGEDGSKKKKNKDKKEKKKEE 509
Score = 52.4 bits (124), Expect = 5e-07
Identities = 35/121 (28%), Positives = 64/121 (51%), Gaps = 17/121 (14%)
Frame = +3
Query: 26 QETEVKGEAE-----------AEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEK 74
+E VKGE E +EK+ +ED+D E KS+ + ++KDK D E +
Sbjct: 276 KEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDEGE 455
Query: 75 EEVEKETDEVKDEVKGEEEKDEE------DEDGGEGEGEGEGEEEEEQEEEEEAEAEADE 128
+ +K+ ++ K E K EE++ EE D D E EG+ ++++++++EE+ + +
Sbjct: 456 DGSKKKKNKDKKEKKKEEDEKEEGKVSVRDIDIEETAKEGKEKKKKKEDKEEKKKKSGKD 635
Query: 129 K 129
K
Sbjct: 636 K 638
Score = 49.3 bits (116), Expect = 4e-06
Identities = 35/126 (27%), Positives = 56/126 (43%), Gaps = 4/126 (3%)
Frame = +3
Query: 383 DDESKSEEEEDKSKAQKKTSKKIVKEGSGSKAGERTKSAKKTPV---KSSKNVDKTPKKS 439
DD+S + +EDK + +K K V++ K + + KT V K K+ +K K+
Sbjct: 96 DDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKKDKEKKKKEK 275
Query: 440 TPKQTAAEHDSASASLSKSKQPASKKRKTENEK-PDTKGKASSKKQTDKSSKALVKDQGK 498
+ E + K K+ KK K + +K D + K S K + K K D+G+
Sbjct: 276 KEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDEGE 455
Query: 499 SKSGKK 504
S KK
Sbjct: 456 DGSKKK 473
Score = 32.0 bits (71), Expect = 0.68
Identities = 20/81 (24%), Positives = 41/81 (49%)
Frame = +3
Query: 7 EDHNKPPVQAEGDDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDK 66
+D K + E DD + + + + + ++KE +E+ S + E E A+E +
Sbjct: 405 KDKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDIDIE-ETAKEGKE 581
Query: 67 GGDQVEEKEEVEKETDEVKDE 87
+ E+KEE +K++ + KD+
Sbjct: 582 KKKKKEDKEEKKKKSGKDKDQ 644
>TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown protein
{Arabidopsis thaliana}, partial (17%)
Length = 1237
Score = 62.4 bits (150), Expect = 5e-10
Identities = 81/330 (24%), Positives = 132/330 (39%), Gaps = 55/330 (16%)
Frame = +2
Query: 311 EKKVKKRTRKVTPSKS--PGEASTETPAKKQKKTSESGKKRK---------------GSS 353
E+ V+++ +K + SKS P + S K+ +KT++S +K G+S
Sbjct: 158 ERSVRRKGKKASSSKSTKPSKKSNVVSEKEAEKTADSKSSKKEVPISLNEDSVVEATGTS 337
Query: 354 DVEEDEKAELSDAKDESQENVAAPH---KESDDDESKSE--------EEEDKSKAQKKTS 402
+ +++ KA++S K E+ AA ES+ DE++S+ + K A + S
Sbjct: 338 ENDKEIKAKISSPKAGGLESDAAGSPSPSESNHDENRSKKRVRTKKNDSSAKEVAAEDIS 517
Query: 403 KKIVKEGSGSKAGERTKSAKKTPVKSS------------------KNVDKTPK------- 437
KK+ + S SK SAKK P++SS K DK K
Sbjct: 518 KKVSEGTSDSKVKPARPSAKKGPIRSSDVKTVVHAVMADVGSSSLKPEDKKKKTHVKGSS 697
Query: 438 -KSTPKQTAAEHDSASASLSKSKQPASKKRKTENEKPDTKGKASSKKQTDKSSKALVKDQ 496
K K +A + D + S KS A+K K E+ + K K+ K + K
Sbjct: 698 EKGLAKSSAEDEDKVTVSSLKS---ATKTTKDEHSEETPKTTLKRKRTPGKEKGSDTKKN 868
Query: 497 GKSKSGKKAKA-VPSREAMHAVVVDILKEVDFNTATLSDILRQLGTHFGVDLMHRKAEVK 555
+S GK+ K P + VVD F+++T + +
Sbjct: 869 DQSLVGKRVKVWWPDDNMFYKGVVD-----SFDSST-----------------KKHKVLY 982
Query: 556 DIITDVINNMSDEEDEGEEADGDGDADKDD 585
D + I N +E+ E E D D D D ++
Sbjct: 983 DDGDEEILNFKEEKYEIVEVDADADPDVEE 1072
>TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I3.230 -
Arabidopsis thaliana, partial (14%)
Length = 1074
Score = 61.2 bits (147), Expect = 1e-09
Identities = 47/169 (27%), Positives = 80/169 (46%), Gaps = 3/169 (1%)
Frame = +3
Query: 307 LADNE-KKVKKRTRKVTPSKSPGEASTETPAKKQKKTSESGKKRKGSSDVEEDEKAELSD 365
+AD E +KVKK + K A + PAKK K G+++ + V +D E+ D
Sbjct: 204 VADEEGEKVKKDDGEGRKRKKHESADSPAPAKKSKVAEVDGEEKVKTKKV-DDAAVEVED 380
Query: 366 AKDESQENVAAPHKESDDDESKSEEEEDKSKAQKKTSKKIVKEGSG--SKAGERTKSAKK 423
K E ++ K+ ++ + + +EE K +KK K+ ++GS K+ ++ K K+
Sbjct: 381 DKKEKKKK----KKKDKENGAAASDEEKVEKEKKKKHKEKGEDGSPEVEKSDKKKKKHKE 548
Query: 424 TPVKSSKNVDKTPKKSTPKQTAAEHDSASASLSKSKQPASKKRKTENEK 472
T S VDK+ KK K A+ ++A S + A + K +K
Sbjct: 549 TSEVGSPEVDKSEKKKKKKDKEAKDNAADISNGNDESNADRSEKKHKKK 695
Score = 45.1 bits (105), Expect = 8e-05
Identities = 38/142 (26%), Positives = 66/142 (45%), Gaps = 1/142 (0%)
Frame = +3
Query: 16 AEGDDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDK-GGDQVEEK 74
A+ A ++ EV GE + + K+ +D E K E + ++KDK G ++
Sbjct: 276 ADSPAPAKKSKVAEVDGEEKVKTKKV---DDAAVEVEDDKKEKKKKKKKDKENGAAASDE 446
Query: 75 EEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKR 134
E+VEK E+K + E G +G E E ++++++ +E +E + E K+++
Sbjct: 447 EKVEK------------EKKKKHKEKGEDGSPEVEKSDKKKKKHKETSEVGSPEVDKSEK 590
Query: 135 GPKKGGKKETASEKKDPATPAS 156
KK K E KD A S
Sbjct: 591 KKKKKDK-----EAKDNAADIS 641
Score = 43.9 bits (102), Expect = 2e-04
Identities = 34/144 (23%), Positives = 64/144 (43%)
Frame = +3
Query: 20 DAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEK 79
+ A E K + E +++ D A A+ K + EEK K + EVE
Sbjct: 201 EVADEEGEKVKKDDGEGRKRKKHESADSPAPAKKSKVAEVDGEEKVKTKKVDDAAVEVED 380
Query: 80 ETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGPKKG 139
+ E K ++K ++ E+G E + E+E++++ +E+ E D + ++ KK
Sbjct: 381 DKKEKK------KKKKKDKENGAAASDEEKVEKEKKKKHKEKGE---DGSPEVEKSDKKK 533
Query: 140 GKKETASEKKDPATPASERPTRER 163
K + SE P SE+ +++
Sbjct: 534 KKHKETSEVGSPEVDKSEKKKKKK 605
Score = 42.0 bits (97), Expect = 7e-04
Identities = 48/213 (22%), Positives = 88/213 (40%), Gaps = 13/213 (6%)
Frame = +3
Query: 67 GGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEE---QEEEEEAE 123
GGD V T++VK EV +EE ++ +D GEG + E + ++ + AE
Sbjct: 141 GGDAVIAGMAAATGTEDVKKEV-ADEEGEKVKKDDGEGRKRKKHESADSPAPAKKSKVAE 317
Query: 124 AEADEKVKAKR---------GPKKGGKKETASEKKDPATPA-SERPTRERKTVERYSVPS 173
+ +EKVK K+ KK KK+ +K++ A + E+ +E+K +
Sbjct: 318 VDGEEKVKTKKVDDAAVEVEDDKKEKKKKKKKDKENGAAASDEEKVEKEKKKKHKEKGED 497
Query: 174 PSKSAKSSGGKGLVIEKGRGEQLKDIPNVAFKLSKRKPDDNLHALHSILFGKRTKAYNLK 233
S + S K K + ++ ++ + S++K K+ K K
Sbjct: 498 GSPEVEKSDKK-----KKKHKETSEVGSPEVDKSEKK--------------KKKKDKEAK 620
Query: 234 RNIGQFSGFVWTENEEKQRAKVKERIDKYVKEK 266
N S N ++ K K++ +K +E+
Sbjct: 621 DNAADISNGNDESNADRSEKKHKKKKNKDAQEE 719
>AL373344 similar to GP|9758600|dbj| gene_id:MDF20.10~pir||T08929~similar to
unknown protein {Arabidopsis thaliana}, partial (2%)
Length = 488
Score = 60.8 bits (146), Expect = 1e-09
Identities = 30/59 (50%), Positives = 41/59 (68%)
Frame = +2
Query: 523 KEVDFNTATLSDILRQLGTHFGVDLMHRKAEVKDIITDVINNMSDEEDEGEEADGDGDA 581
K+VDFNTAT +DIL+ LG F VDL RKA VK II + +++E++E E+ + GDA
Sbjct: 2 KKVDFNTATFADILKLLGKQFDVDLTPRKASVKTIIQQELAKLAEEDEEREQRERRGDA 178
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 60.5 bits (145), Expect = 2e-09
Identities = 31/101 (30%), Positives = 56/101 (54%)
Frame = +3
Query: 34 AEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEE 93
A E K +++D + V+ E ++ EE+D GD+ EE+ + E D+ + G +
Sbjct: 297 AYKENKSDTEDDEDDDDDDDVQDEDDDGEEEDYSGDEGEEEGDPE---DDPEANGAGGSD 467
Query: 94 KDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKR 134
E+D+D G+ E E +GE+EE++E+EEE + K ++
Sbjct: 468 DGEDDDDDGDEEDEEDGEDEEDEEDEEEDDETPQPPAKKRK 590
Score = 43.9 bits (102), Expect = 2e-04
Identities = 24/92 (26%), Positives = 47/92 (51%)
Frame = +3
Query: 73 EKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKA 132
E +E + + D+V+DE +++ +EED G EGE EG+ E++ E ++ D+
Sbjct: 324 EDDEDDDDDDDVQDE---DDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDG 494
Query: 133 KRGPKKGGKKETASEKKDPATPASERPTRERK 164
++ G+ E E ++ + P ++RK
Sbjct: 495 DEEDEEDGEDEEDEEDEEEDDETPQPPAKKRK 590
Score = 37.7 bits (86), Expect = 0.012
Identities = 21/89 (23%), Positives = 40/89 (44%)
Frame = +3
Query: 91 EEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGPKKGGKKETASEKKD 150
++E D++D+D + + +GE E+ E EEE + E D + G G + +++D
Sbjct: 327 DDEDDDDDDDVQDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGDEED 506
Query: 151 PATPASERPTRERKTVERYSVPSPSKSAK 179
E + + + P P+K K
Sbjct: 507 EEDGEDEEDEEDEEEDDETPQP-PAKKRK 590
Score = 32.0 bits (71), Expect = 0.68
Identities = 12/28 (42%), Positives = 20/28 (70%)
Frame = +3
Query: 564 NMSDEEDEGEEADGDGDADKDDNGDDDD 591
N SD ED+ ++ D D D+DD+G+++D
Sbjct: 309 NKSDTEDDEDDDDDDDVQDEDDDGEEED 392
Score = 32.0 bits (71), Expect = 0.68
Identities = 16/64 (25%), Positives = 30/64 (46%), Gaps = 4/64 (6%)
Frame = +3
Query: 345 SGKKRKGSSDVEEDEKAE----LSDAKDESQENVAAPHKESDDDESKSEEEEDKSKAQKK 400
SG + + D E+D +A D +D+ + ++ +D+E + +EEED Q
Sbjct: 396 SGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGDEEDEEDGEDEEDEEDEEEDDETPQPP 575
Query: 401 TSKK 404
K+
Sbjct: 576 AKKR 587
Score = 31.2 bits (69), Expect = 1.2
Identities = 11/29 (37%), Positives = 20/29 (68%)
Frame = +3
Query: 563 NNMSDEEDEGEEADGDGDADKDDNGDDDD 591
+++ DE+D+GEE D GD +++ +DD
Sbjct: 351 DDVQDEDDDGEEEDYSGDEGEEEGDPEDD 437
Score = 30.8 bits (68), Expect = 1.5
Identities = 10/29 (34%), Positives = 21/29 (71%)
Frame = +3
Query: 564 NMSDEEDEGEEADGDGDADKDDNGDDDDE 592
N + D+GE+ D DGD + +++G+D+++
Sbjct: 447 NGAGGSDDGEDDDDDGDEEDEEDGEDEED 533
Score = 29.3 bits (64), Expect = 4.4
Identities = 10/25 (40%), Positives = 18/25 (72%)
Frame = +3
Query: 567 DEEDEGEEADGDGDADKDDNGDDDD 591
D+ DE +E DG+ + D++D +DD+
Sbjct: 486 DDGDEEDEEDGEDEEDEEDEEEDDE 560
>AJ500818 weakly similar to GP|10435304|dbj unnamed protein product {Homo
sapiens}, partial (12%)
Length = 457
Score = 60.1 bits (144), Expect = 2e-09
Identities = 33/106 (31%), Positives = 60/106 (56%)
Frame = -3
Query: 44 QEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGE 103
+ D +A A K +A+ EEK + + EEK + EK +E + E K +EEK +E++ E
Sbjct: 320 KRDAEAEAYGYKRDADPVEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEE 141
Query: 104 GEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGPKKGGKKETASEKK 149
E + +EE+ QEE+ + E +EK + ++ ++ ++E E+K
Sbjct: 140 KRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRLEEKRQEEKRQEEK 3
Score = 54.7 bits (130), Expect = 1e-07
Identities = 38/118 (32%), Positives = 60/118 (50%), Gaps = 4/118 (3%)
Frame = -3
Query: 16 AEGDDAAPNNQETEVKGEAEAE----EKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQV 71
AE D K +AEAE +++A P E++ E + E + EEK + +
Sbjct: 365 AEADAYGYKRDAYGYKRDAEAEAYGYKRDADPVEEK--RQEEKRQEEKRQEEKRQEEKRQ 192
Query: 72 EEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEK 129
EEK + EK +E + E K +EEK +E++ E E + +EE+ QEE+ E +EK
Sbjct: 191 EEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRLEEKRQEEK 18
Score = 51.6 bits (122), Expect = 8e-07
Identities = 29/87 (33%), Positives = 46/87 (52%)
Frame = -3
Query: 33 EAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEE 92
E EEK + E+ E + E + EEK + + EEK + EK +E + E K +E
Sbjct: 263 EKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQE 84
Query: 93 EKDEEDEDGGEGEGEGEGEEEEEQEEE 119
EK +E++ E E + +EE+ QEE+
Sbjct: 83 EKRQEEKRQEEKRLEEKRQEEKRQEEK 3
Score = 51.6 bits (122), Expect = 8e-07
Identities = 29/87 (33%), Positives = 46/87 (52%)
Frame = -3
Query: 37 EEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDE 96
EEK + E+ E + E + EEK + + EEK + EK +E + E K +EEK +
Sbjct: 266 EEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQ 87
Query: 97 EDEDGGEGEGEGEGEEEEEQEEEEEAE 123
E++ E E + EE+ QEE+ + E
Sbjct: 86 EEKRQEEKRQEEKRLEEKRQEEKRQEE 6
Score = 49.7 bits (117), Expect = 3e-06
Identities = 38/134 (28%), Positives = 65/134 (48%), Gaps = 9/134 (6%)
Frame = -3
Query: 44 QEDEDASAEAVKSEA----ENAEEKDKG----GDQVEEKEEVEKETDEVKDEVKGEEEKD 95
+ D +A A K +A +AE + G D VEEK + EK +E + E K +EEK
Sbjct: 374 KRDAEADAYGYKRDAYGYKRDAEAEAYGYKRDADPVEEKRQEEKRQEEKRQEEKRQEEKR 195
Query: 96 EEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEK-VKAKRGPKKGGKKETASEKKDPATP 154
+E++ +EE+ QEE+ + E +EK + KR +K +++ EK+
Sbjct: 194 QEEKR----------QEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKR 45
Query: 155 ASERPTRERKTVER 168
E+ E++ E+
Sbjct: 44 LEEKRQEEKRQEEK 3
Score = 43.1 bits (100), Expect = 3e-04
Identities = 24/74 (32%), Positives = 39/74 (52%)
Frame = -3
Query: 26 QETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVK 85
QE + + E EEK + E+ E + E + EEK + + EEK + EK +E +
Sbjct: 224 QEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKR 45
Query: 86 DEVKGEEEKDEEDE 99
E K +EEK +E++
Sbjct: 44 LEEKRQEEKRQEEK 3
Score = 42.4 bits (98), Expect = 5e-04
Identities = 25/91 (27%), Positives = 42/91 (45%)
Frame = -3
Query: 4 ETLEDHNKPPVQAEGDDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEE 63
+ +E+ + + E QE + + E EEK + E+ E + E + EE
Sbjct: 275 DPVEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEE 96
Query: 64 KDKGGDQVEEKEEVEKETDEVKDEVKGEEEK 94
K + + EEK + EK +E + E K +EEK
Sbjct: 95 KRQEEKRQEEKRQEEKRLEEKRQEEKRQEEK 3
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 59.7 bits (143), Expect = 3e-09
Identities = 39/126 (30%), Positives = 64/126 (49%)
Frame = +3
Query: 16 AEGDDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKE 75
AEG+D + E + G+ E E ++ + N+++ +GG + E
Sbjct: 411 AEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKKAPEGGAGGAD-E 587
Query: 76 EVEKETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRG 135
E+E DE D+ ++++DE+D+D EG GE +EEE +EE+ E E DE +A +
Sbjct: 588 NGEEEDDEDGDDQDEDDDEDEDDDDEEEG---GEEDEEEGVDEEDNEEEEEDEDEEALQP 758
Query: 136 PKKGGK 141
PKK K
Sbjct: 759 PKKRKK 776
Score = 44.3 bits (103), Expect = 1e-04
Identities = 24/85 (28%), Positives = 45/85 (52%), Gaps = 3/85 (3%)
Frame = +3
Query: 69 DQVEEKEEVEKETDEVKDEVKGEEEKDEEDEDGGEGEGE-GEGEEEEEQEEEEEAEAEAD 127
D K + + ++ + + + +GE+ D+EDE+ +G+G GEGE+E E+ ++
Sbjct: 354 DMPTTKGDSKTQSQDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSN 533
Query: 128 EKVKAKRGPK--KGGKKETASEKKD 150
K +K+ P+ GG E E+ D
Sbjct: 534 NKSNSKKAPEGGAGGADENGEEEDD 608
Score = 34.3 bits (77), Expect = 0.14
Identities = 19/84 (22%), Positives = 38/84 (44%)
Frame = +3
Query: 320 KVTPSKSPGEASTETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHK 379
K K+P + +++ E G + D +ED+ E +++ +E V
Sbjct: 537 KSNSKKAPEGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGV----D 704
Query: 380 ESDDDESKSEEEEDKSKAQKKTSK 403
E D++E + +E+E+ + KK K
Sbjct: 705 EEDNEEEEEDEDEEALQPPKKRKK 776
Score = 34.3 bits (77), Expect = 0.14
Identities = 18/74 (24%), Positives = 33/74 (44%)
Frame = +3
Query: 331 STETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHKESDDDESKSEE 390
S + P E+G++ ++DE + + D+ +E +E D+E EE
Sbjct: 546 SKKAPEGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEE 725
Query: 391 EEDKSKAQKKTSKK 404
EED+ + + KK
Sbjct: 726 EEDEDEEALQPPKK 767
Score = 31.6 bits (70), Expect = 0.89
Identities = 23/96 (23%), Positives = 40/96 (40%)
Frame = +3
Query: 328 GEASTETPAKKQKKTSESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHKESDDDESK 387
G + KK E G G+ + E+E E D +DE + ++ DDD+ +
Sbjct: 513 GYGNNSNNKSNSKKAPEGGAG--GADENGEEEDDEDGDDQDEDDD------EDEDDDDEE 668
Query: 388 SEEEEDKSKAQKKTSKKIVKEGSGSKAGERTKSAKK 423
EED+ + + + +E +A + K KK
Sbjct: 669 EGGEEDEEEGVDEEDNEEEEEDEDEEALQPPKKRKK 776
Score = 29.3 bits (64), Expect = 4.4
Identities = 11/26 (42%), Positives = 19/26 (72%)
Frame = +3
Query: 567 DEEDEGEEADGDGDADKDDNGDDDDE 592
+EED+ + D D D D+D++ DD++E
Sbjct: 594 EEEDDEDGDDQDEDDDEDEDDDDEEE 671
Score = 28.9 bits (63), Expect = 5.8
Identities = 11/27 (40%), Positives = 18/27 (65%)
Frame = +3
Query: 566 SDEEDEGEEADGDGDADKDDNGDDDDE 592
+DE E E+ + D D+DD+ D+DD+
Sbjct: 579 ADENGEEEDDEDGDDQDEDDDEDEDDD 659
>TC88579 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (40%)
Length = 1069
Score = 58.2 bits (139), Expect = 9e-09
Identities = 47/152 (30%), Positives = 77/152 (49%), Gaps = 5/152 (3%)
Frame = +2
Query: 341 KTSESGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHKESDDDESKSEEEEDKSKAQKK 400
K + SGK ++ EE+ +++ S + E ESD +ES+SEEE S+++++
Sbjct: 416 KAANSGKP----AEPEEESESDFSGSDISINE-------ESDTEESESEEETPASESEEE 562
Query: 401 TSKKIVKEGSGSKAGERTKSAKKTPVKSSKNVDKTPKKSTPKQT---AAEHDSASASLSK 457
T K V EG K T S +TPV + K D TP+K+ K++ A H K
Sbjct: 563 TPAKKVDEGKNKKRTNETGS--QTPVPTKKAKDATPEKTDGKKSVHIATPHPMKKG--GK 730
Query: 458 SKQPASKKRKTENEKPDT--KGKASSKKQTDK 487
+ Q A+K + ++KP T G+ + K + D+
Sbjct: 731 TPQNAAKDQSPISKKPATTKSGQQNKKSKQDR 826
Score = 34.7 bits (78), Expect = 0.11
Identities = 31/97 (31%), Positives = 42/97 (42%), Gaps = 4/97 (4%)
Frame = +2
Query: 92 EEKDEEDEDGGE-GEGEGEGEEEEEQEEEEEAEAEADEKVKAKR---GPKKGGKKETASE 147
EE+ E D G + E EE E EEE A +E++E+ AK+ G K ET S+
Sbjct: 449 EEESESDFSGSDISINEESDTEESESEEETPA-SESEEETPAKKVDEGKNKKRTNETGSQ 625
Query: 148 KKDPATPASERPTRERKTVERYSVPSPSKSAKSSGGK 184
P A + KT + SV + GGK
Sbjct: 626 TPVPTKKAKD--ATPEKTDGKKSVHIATPHPMKKGGK 730
>TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsis
thaliana}, partial (32%)
Length = 1558
Score = 57.4 bits (137), Expect = 2e-08
Identities = 38/139 (27%), Positives = 73/139 (52%), Gaps = 13/139 (9%)
Frame = +3
Query: 39 KEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVEKETDEVKDEVKGEEEKDEED 98
KE A +++ E K E E EEK+K +EKE+ EKE + K++ K +KDE D
Sbjct: 720 KEKAKEKERKREEEKAKKEKER-EEKEKR----KEKEKKEKEREREKEKSKERHKKDESD 884
Query: 99 EDGGE-GEGEGEGEEEEEQEEEE------------EAEAEADEKVKAKRGPKKGGKKETA 145
D + +G G EE+++++++E + ++E DEK ++++ + G ++
Sbjct: 885 SDNQDMTDGHGYREEKKKEKDKERKHRRRHQSSMDDVDSEKDEKEESRKSRRHGSDRK-- 1058
Query: 146 SEKKDPATPASERPTRERK 164
+K +P S+ +R ++
Sbjct: 1059KSRKHANSPESDNESRHKR 1115
Score = 38.5 bits (88), Expect = 0.007
Identities = 27/118 (22%), Positives = 55/118 (45%), Gaps = 3/118 (2%)
Frame = +3
Query: 26 QETEVKGEAEAEEKEAAPQEDEDASAEAVK---SEAENAEEKDKGGDQVEEKEEVEKETD 82
+E E + E E +EKE + +++ S E K S+++N + D G + E+K+E +KE
Sbjct: 786 EEKEKRKEKEKKEKER--EREKEKSKERHKKDESDSDNQDMTDGHGYREEKKKEKDKERK 959
Query: 83 EVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEKVKAKRGPKKGG 140
+ ++ D E ++ E + ++ + E+D + + KR + G
Sbjct: 960 HRRRHQSSMDDVDSEKDEKEESRKSRRHGSDRKKSRKHANSPESDNESRHKRHKRDHG 1133
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 56.2 bits (134), Expect = 3e-08
Identities = 27/111 (24%), Positives = 61/111 (54%)
Frame = +1
Query: 19 DDAAPNNQETEVKGEAEAEEKEAAPQEDEDASAEAVKSEAENAEEKDKGGDQVEEKEEVE 78
D P +Q ++ E + + ++D+D + ++ ++ ++D GD E+ E+ +
Sbjct: 235 DGRFPLDQLSKGNHTCEENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSGD--EDDEDAD 408
Query: 79 KETDEVKDEVKGEEEKDEEDEDGGEGEGEGEGEEEEEQEEEEEAEAEADEK 129
E D V + G ++ DE+D+D + +GE E+E+E+E+++E + + +K
Sbjct: 409 PEDDPVPNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDDEDQPPSKKK 561
Score = 32.7 bits (73), Expect = 0.40
Identities = 13/26 (50%), Positives = 19/26 (73%)
Frame = +1
Query: 567 DEEDEGEEADGDGDADKDDNGDDDDE 592
DE+D+ + D D D D+D +GD+DDE
Sbjct: 322 DEDDDDDVNDEDDDNDEDFSGDEDDE 399
Score = 30.8 bits (68), Expect = 1.5
Identities = 16/60 (26%), Positives = 28/60 (46%)
Frame = +1
Query: 345 SGKKRKGSSDVEEDEKAELSDAKDESQENVAAPHKESDDDESKSEEEEDKSKAQKKTSKK 404
SG + +D E+D + D+ E+ ++DD E + E+EE+ + SKK
Sbjct: 379 SGDEDDEDADPEDDPVPNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDDEDQPPSKK 558
Score = 30.4 bits (67), Expect = 2.0
Identities = 11/30 (36%), Positives = 19/30 (62%)
Frame = +1
Query: 563 NNMSDEEDEGEEADGDGDADKDDNGDDDDE 592
N+ +E +G E + D D D DD+ +D+D+
Sbjct: 271 NHTCEENKDGSETEDDDDEDDDDDVNDEDD 360
Score = 29.6 bits (65), Expect = 3.4
Identities = 14/33 (42%), Positives = 20/33 (60%)
Frame = +1
Query: 560 DVINNMSDEEDEGEEADGDGDADKDDNGDDDDE 592
D + N + D+ +E D D D D DD G+D+DE
Sbjct: 418 DPVPNGAGGSDDDDEDDDDDDDDNDD-GEDEDE 513
Score = 28.9 bits (63), Expect = 5.8
Identities = 9/26 (34%), Positives = 19/26 (72%)
Frame = +1
Query: 567 DEEDEGEEADGDGDADKDDNGDDDDE 592
DE+D+ ++ D D D+D++ ++DD+
Sbjct: 457 DEDDDDDDDDNDDGEDEDEDEEEDDD 534
Score = 28.1 bits (61), Expect = 9.9
Identities = 11/25 (44%), Positives = 17/25 (68%)
Frame = +1
Query: 566 SDEEDEGEEADGDGDADKDDNGDDD 590
S+ ED+ +E D D D+DD+ D+D
Sbjct: 301 SETEDDDDEDDDDDVNDEDDDNDED 375
Score = 28.1 bits (61), Expect = 9.9
Identities = 9/26 (34%), Positives = 18/26 (68%)
Frame = +1
Query: 567 DEEDEGEEADGDGDADKDDNGDDDDE 592
D++D+ ++ D D D+D+ DDD++
Sbjct: 463 DDDDDDDDNDDGEDEDEDEEEDDDED 540
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.296 0.120 0.306
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,425,252
Number of Sequences: 36976
Number of extensions: 133781
Number of successful extensions: 4318
Number of sequences better than 10.0: 549
Number of HSP's better than 10.0 without gapping: 1977
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3308
length of query: 592
length of database: 9,014,727
effective HSP length: 102
effective length of query: 490
effective length of database: 5,243,175
effective search space: 2569155750
effective search space used: 2569155750
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0063.6


