

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.10
(306 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
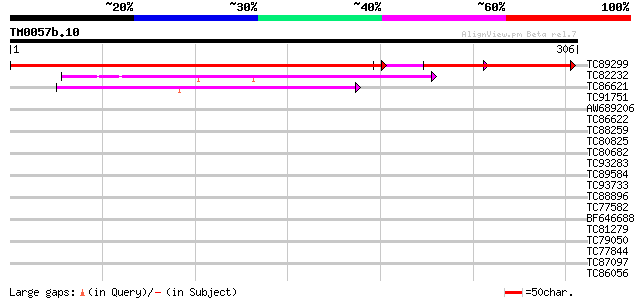
Score E
Sequences producing significant alignments: (bits) Value
TC89299 similar to GP|17979458|gb|AAL50066.1 At2g30880/F7F1.9 {A... 294 2e-97
TC82232 similar to GP|10177316|dbj|BAB10642. kinesin-like protei... 45 3e-05
TC86621 similar to PIR|F84664|F84664 hypothetical protein At2g26... 44 1e-04
TC91751 similar to GP|2739382|gb|AAC14505.1| unknown protein {Ar... 39 0.003
AW689206 similar to GP|17473880|gb| putative protein {Arabidopsi... 39 0.003
TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myo... 38 0.004
TC88259 similar to GP|6729037|gb|AAF27033.1| unknown protein {Ar... 37 0.012
TC80825 similar to PIR|F84614|F84614 probable kinesin heavy chai... 36 0.021
TC80682 similar to GP|10086489|gb|AAG12549.1 Unknown Protein {Ar... 35 0.028
TC93283 weakly similar to GP|23476966|emb|CAD43403. SMC3 protein... 35 0.028
TC89584 weakly similar to GP|18076679|emb|CAC84774. P70 protein ... 35 0.028
TC93733 similar to GP|10998143|dbj|BAB03114. kinesin (centromere... 35 0.028
TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown prot... 35 0.028
TC77582 similar to SP|Q43207|FKB7_WHEAT 70 kDa peptidylprolyl is... 35 0.036
BF646688 similar to GP|3851530|gb| nodulin {Glycine max}, partia... 35 0.036
TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.... 34 0.062
TC79050 homologue to GP|18030195|gb|AAG00027.2 Hypothetical prot... 34 0.062
TC77844 similar to PIR|T49049|T49049 hypothetical protein T5P19.... 33 0.18
TC87097 similar to GP|14334540|gb|AAK59678.1 putative protein {A... 32 0.23
TC86056 similar to GP|8777369|dbj|BAA96959.1 gene_id:MJE7.2~pir|... 32 0.23
>TC89299 similar to GP|17979458|gb|AAL50066.1 At2g30880/F7F1.9 {Arabidopsis
thaliana}, partial (85%)
Length = 1744
Score = 294 bits (752), Expect(2) = 2e-97
Identities = 156/203 (76%), Positives = 175/203 (85%)
Frame = +2
Query: 1 MQISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLS 60
MQISLRN LGMM N+TTDGPMDDLTIMKETL VKDEELQN ARDLR RDSTI +IA+KLS
Sbjct: 629 MQISLRNALGMMPNRTTDGPMDDLTIMKETLRVKDEELQNLARDLRARDSTITEIADKLS 808
Query: 61 ETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKE 120
ETAEAAEAAASAA+T+DEQ R C EIE L+KDSEKQ ++ AQKLKEY+ KI LSKE+E
Sbjct: 809 ETAEAAEAAASAAYTMDEQRRIVCAEIERLRKDSEKQQQVYAQKLKEYKEKITCLSKERE 988
Query: 121 QLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGES 180
QL+ QR+AA +EANMWR+ELGKARE VILE + RAEEKVRVAEANAEA+IKE+VQ ES
Sbjct: 989 QLTDQRDAAIKEANMWRTELGKAREHDVILEATVVRAEEKVRVAEANAEARIKESVQRES 1168
Query: 181 AAVSEKQELLAYVDKLKAQLQRQ 203
A EKQELLAYV+ LKAQLQR+
Sbjct: 1169VATKEKQELLAYVNMLKAQLQRR 1237
Score = 79.7 bits (195), Expect(2) = 2e-97
Identities = 47/83 (56%), Positives = 53/83 (63%), Gaps = 1/83 (1%)
Frame = +1
Query: 224 DLTEENVDKACLSVSRSVPDPAAAECVVQMPTDQVIIQPVGDNEWSDIQATEARVADVRE 283
D TEENVDK V T IQPVGDNEWSDIQATEAR++DVRE
Sbjct: 1300 DPTEENVDKGMPKVFLEQTLSLQRM*FTWQQTR*TNIQPVGDNEWSDIQATEARISDVRE 1479
Query: 284 VAS-ESEVSSLDIPVISQQGTNH 305
VA+ E++ SSLDIPV+SQ G NH
Sbjct: 1480 VATPETDGSSLDIPVVSQPGINH 1548
Score = 48.9 bits (115), Expect = 2e-06
Identities = 32/63 (50%), Positives = 38/63 (59%), Gaps = 1/63 (1%)
Frame = +3
Query: 197 KAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVD-KACLSVSRSVPDPAAAECVVQMPT 255
K + +HID+T+V EKTESCSDTKHV + VSR+ P P AE VV M T
Sbjct: 1218 KHNFKGEHIDATQVVEKTESCSDTKHV*SH*RKCG*RHA*GVSRANPIP--AENVVHMAT 1391
Query: 256 DQV 258
DQV
Sbjct: 1392 DQV 1400
>TC82232 similar to GP|10177316|dbj|BAB10642. kinesin-like protein
{Arabidopsis thaliana}, partial (21%)
Length = 1181
Score = 45.1 bits (105), Expect = 3e-05
Identities = 43/220 (19%), Positives = 102/220 (45%), Gaps = 18/220 (8%)
Frame = +3
Query: 29 ETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIE 88
E + + ++EL++ + + D ++++ +KL E EA + A + + + E+E
Sbjct: 153 EPVEIHEKELEHSSAQQKL-DRELKELDKKL-EQKEAEMKLVNNASVLKQHYEKKLNELE 326
Query: 89 SLKKDSEKQLEL-------SAQKLKE-YEVKIISLSKEKEQLSKQREAAT---------- 130
KK ++++E S KLKE Y K+ +L + +L K+++A
Sbjct: 327 HEKKFLQREIEELKSTSGDSTHKLKEEYLQKLNALESQVSELKKKQDAQAHLLRQKQKGD 506
Query: 131 QEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELL 190
+ A + E+ + + V L+ + + E+ R+ +A+ E ++ + + E+ +LL
Sbjct: 507 EAAKRLQDEIQRIKSQKVQLQHKIKQESEQFRLWKASREKEVLQLKKEGRKNEYERHKLL 686
Query: 191 AYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENV 230
A + K LQR+ +++ ++ + +++ E +
Sbjct: 687 ALNQRTKMVLQRKTEEASLATKRLKELLESRKASSRETGI 806
>TC86621 similar to PIR|F84664|F84664 hypothetical protein At2g26770
[imported] - Arabidopsis thaliana, partial (97%)
Length = 1890
Score = 43.5 bits (101), Expect = 1e-04
Identities = 41/173 (23%), Positives = 81/173 (46%), Gaps = 9/173 (5%)
Frame = +1
Query: 26 IMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACE 85
I +ET ++ D+ + RDL ++ A KLSE A EAA+ H + ++ R++ E
Sbjct: 301 IARETAMLLDQHNRLSVRDLASKFEKGLAAAAKLSEEARLREAASLEKHVLLKKLRDSLE 480
Query: 86 EIESL--------KKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEA-NMW 136
++ +D+ +E A +L + E +++ E ++L+ + A+++A +
Sbjct: 481 SLKGRVAGRNKDDVEDAIAMVEALAVQLTQREGELLQEKAEVKKLANFLKQASEDAKKLV 660
Query: 137 RSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQEL 189
E AR A+ R EE ++ E ++A K+ V+ V E + +
Sbjct: 661 DEERAFARAEIDNARSAVQRVEESLQEHERMSQASGKQDVEQLMKEVQEARRI 819
>TC91751 similar to GP|2739382|gb|AAC14505.1| unknown protein {Arabidopsis
thaliana}, partial (38%)
Length = 1200
Score = 38.5 bits (88), Expect = 0.003
Identities = 48/179 (26%), Positives = 81/179 (44%), Gaps = 7/179 (3%)
Frame = +2
Query: 76 IDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANM 135
++++ A EEI +K +E + Q LKE + S + E+L E A E
Sbjct: 281 VEQELDKADEEIPEYRKQAETAEQTKNQVLKELD----STKRLIEELKLNLERAQTEEQQ 448
Query: 136 WR--SELGKAR----ESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQEL 189
R SEL K R E G+ E ++A A+ ++ VA+A A I + AAV E+
Sbjct: 449 ARQDSELAKLRVEEMEQGIADESSVA-AKAQLEVAKARYTAAITDL-----AAVKEE--- 601
Query: 190 LAYVDKLKAQLQRQHIDSTEVFEKT-ESCSDTKHVDLTEENVDKACLSVSRSVPDPAAA 247
+D L+ + D E +K E+ + +K V+ + E++ ++ S+ AA
Sbjct: 602 ---LDALRKEYASLVTDRDEAIKKAEEAVTASKEVEKSVEDLTIELIATKESLETAHAA 769
>AW689206 similar to GP|17473880|gb| putative protein {Arabidopsis thaliana},
partial (6%)
Length = 615
Score = 38.5 bits (88), Expect = 0.003
Identities = 26/137 (18%), Positives = 64/137 (45%), Gaps = 11/137 (8%)
Frame = +2
Query: 26 IMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACE 85
+ +E +D+ + N ++ ++ + +I + +ET +A + + + + +
Sbjct: 80 LQEEEARRQDKLVSNLTNIIQVKNKHLHEIEARCTETTAKMNSAMTEKDQLIQAYNEEIK 259
Query: 86 EIESLKKDSEKQLELSAQKLK--------EYEVKIISLSKEKEQLSKQREAATQEA---N 134
+I+S KD +++ +KLK E E + I+L K + +R +E N
Sbjct: 260 KIQSSAKDHFQRIFNDHEKLKSQLETQKSELESRRITLEKREAHNESERRKLLEEMEELN 439
Query: 135 MWRSELGKARESGVILE 151
+W GK++++G +++
Sbjct: 440 LWMINSGKSQKTGNMIK 490
>TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myosin heavy
chain {Arabidopsis thaliana}, partial (17%)
Length = 2546
Score = 38.1 bits (87), Expect = 0.004
Identities = 52/237 (21%), Positives = 106/237 (43%), Gaps = 26/237 (10%)
Frame = +2
Query: 5 LRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRT---RDSTIRDIAEKLSE 61
L + L ++ K + + L I+++ L + +LQ+ DL+ R+S IR+ + E
Sbjct: 200 LEDALNSLSEKLAESE-NLLEIVRDDLNLTQVKLQSTENDLKAAELRESEIREKHNAIEE 376
Query: 62 T--AEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEK 119
+ ++A ++ + + ESL +DSE++L+ + +K + ++ SL ++
Sbjct: 377 NLAVRGRDIELTSARNLELESLH-----ESLTRDSEQKLQEAIEKFNSKDSEVQSLLEKI 541
Query: 120 EQLSKQREAATQEANMWRSE-------LGKARESGVILEGALARAEEKVRVAEANAE--- 169
+ L + A +++ +SE L + L+ + AE+K + + E
Sbjct: 542 KILEENIAGAGEQSISLKSEFEESLSKLASLQSENEDLKRQIVEAEKKTSQSFSENELLV 721
Query: 170 -------AKIKEAVQGESAAVSEK----QELLAYVDKLKAQLQRQHIDSTEVFEKTE 215
KI E + ++ VSEK QEL+++ L A+L S+E+ E
Sbjct: 722 GTNIQLKTKIDELQESLNSVVSEKEVTAQELVSH-KNLLAELNDVQSKSSEIHSANE 889
Score = 36.2 bits (82), Expect = 0.016
Identities = 55/267 (20%), Positives = 106/267 (39%), Gaps = 8/267 (2%)
Frame = +3
Query: 40 NFARDLRTRDSTIRDIAEKLS-------ETAEAAEAAASAAHTIDEQWRNACEEIESLKK 92
NF +++ +S + D+ KLS ET + A+ +AA + Q EE+++LK
Sbjct: 1176 NFVQEIAVYESKLSDLQSKLSAALVEKDETVKEILASKNAAEDLVTQHN---EEVQTLKS 1346
Query: 93 DSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEG 152
+I S+ ++ L++ + +E LE
Sbjct: 1347 ------------------QISSVIDDRNLLNETNQNLKKE-----------------LES 1421
Query: 153 ALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFE 212
+ EEK++ + N E +K V+ ++EK L + + +++AQL + E
Sbjct: 1422 IILDLEEKLKEHQKN-EDSLKSEVETLKIEIAEKSALQSRLHEIEAQLAKAESRLHEEVG 1598
Query: 213 KTESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECVVQMPTDQVIIQPVGDNEWSDIQ 272
++ + + VDL+ + D +V + AE ++ Q I E ++
Sbjct: 1599 SVQAAASQREVDLSSKFEDYEQKISEINVLNGKVAELEKELHLAQDTIANQKGEESQKLE 1778
Query: 273 ATEARVADVREV-ASESEVSSLDIPVI 298
A V E+ ++E+S L VI
Sbjct: 1779 LEAALKNSVEELETKKNEISLLQKQVI 1859
>TC88259 similar to GP|6729037|gb|AAF27033.1| unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1757
Score = 36.6 bits (83), Expect = 0.012
Identities = 41/134 (30%), Positives = 60/134 (44%), Gaps = 17/134 (12%)
Frame = +3
Query: 85 EEIESLKKDSEKQLELS------AQKLKEYEVK---IISLSKEKEQLSKQREAATQEANM 135
EE+ + KK +E+ +++ A L E K + SK E+ E A E +
Sbjct: 381 EEVSTPKKYNEEGIDVKTLTNELAAALLEISAKEDMVKQHSKVAEEAISGWEKAENEVSS 560
Query: 136 WRSELGKARESGVILE-------GALARAEEKVRVAEANAEAKIKEAVQGES-AAVSEKQ 187
+ +L AR+ LE GAL ++R A E KI EAV S + S +
Sbjct: 561 LKQQLDAARQKNSGLEDRVSHLDGALKECMRQLRQAREVQEQKIHEAVANNSHDSGSRRF 740
Query: 188 ELLAYVDKLKAQLQ 201
EL V +L+AQLQ
Sbjct: 741 ELERKVAELEAQLQ 782
>TC80825 similar to PIR|F84614|F84614 probable kinesin heavy chain
[imported] - Arabidopsis thaliana, partial (16%)
Length = 1771
Score = 35.8 bits (81), Expect = 0.021
Identities = 36/178 (20%), Positives = 69/178 (38%), Gaps = 16/178 (8%)
Frame = +2
Query: 36 EELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSE 95
+ LQN + +D+ +++ EK+ E E S ++ SE
Sbjct: 749 KSLQNIESKAKGKDNIHKNLQEKIKELEGQIELKTS------------------MQNQSE 874
Query: 96 KQLELSAQKLKEYEVKIISLSKEKEQLSK------QREAATQEANMW------RSELGKA 143
KQ+ +KLK E +L + ++L + Q E A + W + +L +
Sbjct: 875 KQVSQLCEKLKGKEETCCTLQHKVKELERKIKEQLQTETANFQQKAWDLEKKLKDQLQGS 1054
Query: 144 RESGVILEGALARAEEKVRVAEANAEAKI----KEAVQGESAAVSEKQELLAYVDKLK 197
L+ + E K++ E N+E+ + KE + + Q+ YV+ +K
Sbjct: 1055ESESSFLKDKIKELERKLKEQEQNSESLLKQ*MKELEEKHKEREQQWQQTHCYVEAVK 1228
>TC80682 similar to GP|10086489|gb|AAG12549.1 Unknown Protein {Arabidopsis
thaliana}, partial (15%)
Length = 1374
Score = 35.4 bits (80), Expect = 0.028
Identities = 26/106 (24%), Positives = 46/106 (42%), Gaps = 2/106 (1%)
Frame = +2
Query: 24 LTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQW-RN 82
L M E+ + Q F +L + +L + + EAA ++EQW RN
Sbjct: 329 LKCMAESYRSLETRAQEFETELNHLRMKTETLENELKDEKRSHEAALQKCKELEEQWQRN 508
Query: 83 ACEEIES-LKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQRE 127
++ ++ E+ L +A+KL E + I L K+ + + Q E
Sbjct: 509 ESSAADNEIQTKKERDLASAAEKLAECQETIYLLGKQLKAMHPQTE 646
>TC93283 weakly similar to GP|23476966|emb|CAD43403. SMC3 protein
{Arabidopsis thaliana}, partial (18%)
Length = 902
Score = 35.4 bits (80), Expect = 0.028
Identities = 44/185 (23%), Positives = 81/185 (43%), Gaps = 12/185 (6%)
Frame = +1
Query: 29 ETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIE 88
+++ VK+EEL+ +++ D I D+ + + +D Q ++ EIE
Sbjct: 406 DSIHVKEEELEKVKFEIQEIDQKINDLVTEQQK--------------VDAQCAHSKSEIE 543
Query: 89 SLKKD---SEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARE 145
LK+D S KQ L ++ L + E + + + EQL + AT++A M
Sbjct: 544 ELKRDIANSNKQKPLFSKALAKKEKSLEDVQNQIEQL--KGSIATKKAEM---------- 687
Query: 146 SGVILEGALARAEEKVRVAEANAEAK-IKE--------AVQGESAAVSEKQELLAYVDKL 196
G L L E+K+ +++ N E K +KE ++ E+ + L + K
Sbjct: 688 -GTELIDHLTPXEKKL-LSDLNPEIKDLKEKLVACKTDRIESEARKAELETNLTTNLXKX 861
Query: 197 KAQLQ 201
K +L+
Sbjct: 862 KQELE 876
>TC89584 weakly similar to GP|18076679|emb|CAC84774. P70 protein {Nicotiana
tabacum}, partial (26%)
Length = 872
Score = 35.4 bits (80), Expect = 0.028
Identities = 37/184 (20%), Positives = 82/184 (44%), Gaps = 3/184 (1%)
Frame = +3
Query: 7 NTLGMMTNKTTDGPMDDLTIMKETLL-VKDEELQNFARDLRTRDSTIRDIAEKLSETAEA 65
N+ +N P D +K T + ++ N +L +R S + + +K E +
Sbjct: 354 NSASSSSNPANKTPKDRSPNVKSTQSPISEKNGTNRVHELESRLSQLEEDLKKAKEQLNS 533
Query: 66 AEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSK-EKEQLSK 124
+E+ A E D++ +L +++L+E + +++ LS E E++ +
Sbjct: 534 SESLRIKAQHEAE--------------DAKNRLLSMSRELEESQRQLLELSNSEDERIQE 671
Query: 125 QREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAE-AKIKEAVQGESAAV 183
R+ + W+SEL ++ + ALA A +++ + E A+ EA + ++A
Sbjct: 672 LRKISQDRDREWQSELEAVQKQHSMDSAALASAMNEIQKLKMQLERARESEATRIDNAES 851
Query: 184 SEKQ 187
++ +
Sbjct: 852 ADAE 863
>TC93733 similar to GP|10998143|dbj|BAB03114. kinesin (centromere protein)
like heavy chain-like protein {Arabidopsis thaliana},
partial (13%)
Length = 747
Score = 35.4 bits (80), Expect = 0.028
Identities = 39/166 (23%), Positives = 72/166 (42%), Gaps = 4/166 (2%)
Frame = +1
Query: 36 EELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSE 95
EEL+ +L + +KL+E + A+ ASAA + + EE+ L +E
Sbjct: 28 EELKQKVEELTASKDQLEVRNQKLAEESSYAKGLASAAAV---ELKALSEEVAKLMNHNE 198
Query: 96 K-QLELSAQKLKEYEVKIISLS---KEKEQLSKQREAATQEANMWRSELGKARESGVILE 151
+ EL+A K + + + + Q+ +R + + EL +++ + E
Sbjct: 199 RLSAELAASKNSPTPRRTSGTAQNGRRESQVRLRRNDQGVSNSDVKRELALSKDRELSYE 378
Query: 152 GALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLK 197
AL ++K E + KI+E+ Q E+ +E + V KLK
Sbjct: 379 AALLEKDQK----EVELQRKIEESKQREAYLENELANMWVLVAKLK 504
>TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown protein
{Arabidopsis thaliana}, partial (27%)
Length = 1097
Score = 35.4 bits (80), Expect = 0.028
Identities = 40/155 (25%), Positives = 68/155 (43%), Gaps = 9/155 (5%)
Frame = +2
Query: 86 EIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARE 145
EIE+LK + EK K Y+ K+ EQL+ + EAA + RS L + R+
Sbjct: 17 EIEALKHELEKA--------KGYDEKLAEKETLIEQLNVESEAAKMAESYARSVLDECRK 172
Query: 146 SGVILEGALARAEEKVRVAEANAEAKIKEAVQGESA----AVSEKQELLAYVDKLKAQLQ 201
LE + A + R A + E K+ ++G++ A SE L + L+ +
Sbjct: 173 KVEELEMKVEEANQLERSASLSLETATKQ-LEGKNELLHDAESEISSLKEKLGMLEMTVG 349
Query: 202 RQHIDSTE-----VFEKTESCSDTKHVDLTEENVD 231
RQ D + + K E+ +K ++ E ++
Sbjct: 350 RQRGDLEDAERCLLAAKEENIEMSKKIESLESEIE 454
>TC77582 similar to SP|Q43207|FKB7_WHEAT 70 kDa peptidylprolyl isomerase (EC
5.2.1.8) (Peptidylprolyl cis-trans isomerase) (PPiase),
partial (92%)
Length = 2130
Score = 35.0 bits (79), Expect = 0.036
Identities = 35/156 (22%), Positives = 66/156 (41%), Gaps = 16/156 (10%)
Frame = +1
Query: 94 SEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVIL--- 150
S+++L + YEV+++S KEKE + + A GK +E G +L
Sbjct: 1165 SQQELAVPPNSTVYYEVELVSYEKEKESWDMNTQEKIEAA-------GKKKEEGNVLFKA 1323
Query: 151 ---EGALARAEEKVRVAEANAEAKIKEAVQGESAAVSE-------KQELLAYVDKLKAQL 200
E A R ++ + E ++ +E ++ ++ K +L Y D K
Sbjct: 1324 GKYERASKRYDKAAKYIEYDSSFSEEEKKLSKTLKIASYLNNAACKLKLKEYKDAEKLCT 1503
Query: 201 QRQHIDSTEV---FEKTESCSDTKHVDLTEENVDKA 233
+ I+ST V + + ++ +DL E ++ KA
Sbjct: 1504 KVLDIESTNVKALYRRAQASMQLTDLDLAEIDIKKA 1611
>BF646688 similar to GP|3851530|gb| nodulin {Glycine max}, partial (46%)
Length = 669
Score = 35.0 bits (79), Expect = 0.036
Identities = 35/110 (31%), Positives = 50/110 (44%)
Frame = +3
Query: 91 KKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVIL 150
K D+E ++ ++ Q+ E E + I + E + QREA EAN SEL K +
Sbjct: 18 KIDAETKV-IAMQRAGESEKQGIKVRTEVKVFENQREAEVAEAN---SELAKKK------ 167
Query: 151 EGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQL 200
A +A + V A A + +QGE EK L +KLKA L
Sbjct: 168 -AAWTKAAQVAEVEAKKAVALREAELQGE----VEKMNALTTTEKLKADL 302
>TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.170 -
Arabidopsis thaliana, partial (31%)
Length = 1205
Score = 34.3 bits (77), Expect = 0.062
Identities = 30/120 (25%), Positives = 55/120 (45%), Gaps = 6/120 (5%)
Frame = +2
Query: 44 DLRTRDSTIRDIAEKLSE---TAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLEL 100
++ T I D+ +KL+ T +A T Q+ E E L+++ + E
Sbjct: 656 EILTSREVIDDLNKKLTNCISTIDAKNIELINLQTALGQYYAEIEAKEHLEEELARAREE 835
Query: 101 SA---QKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARA 157
+A Q LK+ + ++ LS EKE++ + + + + WRS + K E L AL ++
Sbjct: 836 TANLSQLLKDADSRVDILSGEKEEILAKLSQSEKVQSEWRSRVSKLEERNAKLRRALEQS 1015
>TC79050 homologue to GP|18030195|gb|AAG00027.2 Hypothetical protein W01C8.3
{Caenorhabditis elegans}, partial (1%)
Length = 494
Score = 34.3 bits (77), Expect = 0.062
Identities = 16/49 (32%), Positives = 31/49 (62%)
Frame = -2
Query: 31 LLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQ 79
L ++ EEL++ ++R + +RD+ EK ++A+AAE A+ A ++ Q
Sbjct: 241 LTLEMEELRSKGEEMREKIEEMRDVVEKSQDSAKAAEVIAARAADLETQ 95
>TC77844 similar to PIR|T49049|T49049 hypothetical protein T5P19.130 -
Arabidopsis thaliana, partial (70%)
Length = 1969
Score = 32.7 bits (73), Expect = 0.18
Identities = 46/187 (24%), Positives = 79/187 (41%), Gaps = 11/187 (5%)
Frame = +2
Query: 13 TNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASA 72
T + GP I +E + ++ + RDL ++ A KLS A+ E +
Sbjct: 416 TAEDNQGPSLKEVIEQEASNLSEQNNRISVRDLASKFDKNLSAAAKLSNEAKLREVPSLE 595
Query: 73 AHTIDEQWRNACEEIESLKKDSEKQ--------LELSAQKLKEYEVKIISLSKEKEQLSK 124
H + ++ R+A E ++ K+ +E A KL + E ++I E ++L
Sbjct: 596 GHVLLKKLRDALEYLKGRFTGRNKEDVANAISMVEALAVKLTQNEGELIQEKFEVKKLLN 775
Query: 125 QREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEA-NAEAKIKEA--VQGESA 181
+ A+++A +L +S E ARA +R+ EA + K+ EA Q
Sbjct: 776 FLKQASEDA----KKLVNQEKSFACAEIESARA-VVLRIGEALEEQEKVTEASKPQDVDG 940
Query: 182 AVSEKQE 188
V E QE
Sbjct: 941 LVEEVQE 961
>TC87097 similar to GP|14334540|gb|AAK59678.1 putative protein {Arabidopsis
thaliana}, partial (62%)
Length = 1234
Score = 32.3 bits (72), Expect = 0.23
Identities = 30/110 (27%), Positives = 48/110 (43%), Gaps = 2/110 (1%)
Frame = +1
Query: 67 EAAASAAHTIDEQWRNACEEIESLKKDSEKQL-ELSAQKLKEYEVKIISLSKE-KEQLSK 124
EA AS + Q R A + E ++ + + E+ K +E E + L +E K Q ++
Sbjct: 334 EAKASKKKELKRQEREARRQAEEARQSKQDRYSEMRRLKDEEREAQERKLEEEAKAQKAR 513
Query: 125 QREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKE 174
+ EAA E + W+ E V EG L ++K N IK+
Sbjct: 514 EEEAAALEFDKWKGEF------SVDDEGTLEEEQDKTEDLLTNFVEYIKK 645
>TC86056 similar to GP|8777369|dbj|BAA96959.1
gene_id:MJE7.2~pir||C71412~similar to unknown protein
{Arabidopsis thaliana}, partial (25%)
Length = 2085
Score = 32.3 bits (72), Expect = 0.23
Identities = 24/98 (24%), Positives = 45/98 (45%), Gaps = 1/98 (1%)
Frame = +1
Query: 52 IRDIAEKLSETAEA-AEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEV 110
I + K+ + +A AE + A T++ +W+ E + L+K +++ + KE+E
Sbjct: 220 IDSTSSKIQQLQKAFAELESYRAVTLNLKWKELEEHFQGLEKSLKRRFNELEDQEKEFEN 399
Query: 111 KIISLSKEKEQLSKQREAATQEANMWRSELGKARESGV 148
K K +E L KQ A + L + R++ V
Sbjct: 400 K---TRKAREMLEKQEAAVFAKEQALLQRLQRKRDASV 504
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.307 0.122 0.315
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,158,825
Number of Sequences: 36976
Number of extensions: 58267
Number of successful extensions: 312
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 307
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 309
length of query: 306
length of database: 9,014,727
effective HSP length: 96
effective length of query: 210
effective length of database: 5,465,031
effective search space: 1147656510
effective search space used: 1147656510
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0057b.10


