

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0055.8
(509 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
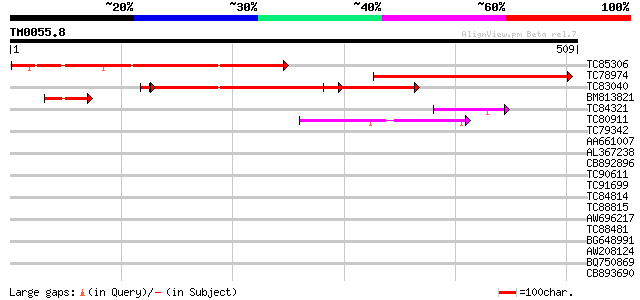
Score E
Sequences producing significant alignments: (bits) Value
TC85306 similar to GP|21593320|gb|AAM65269.1 unknown {Arabidopsi... 328 3e-90
TC78974 similar to GP|22137238|gb|AAM91464.1 AT5g14050/MUA22_5 {... 312 2e-85
TC83040 similar to GP|21593320|gb|AAM65269.1 unknown {Arabidopsi... 250 7e-69
BM813821 PIR|T09297|T09 tonoplast intrinsic protein homolog MSMC... 48 8e-06
TC84321 GP|22138476|gb|AAM93460.1 hypothetical protein {Oryza sa... 44 2e-04
TC80911 similar to GP|3668118|emb|CAA11819.1 hypothetical protei... 42 6e-04
TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein h... 40 0.003
AA661007 similar to GP|21539573|gb| WD40-repeat protein {Arabido... 36 0.031
AL367238 similar to GP|10177204|dbj contains similarity to unkno... 36 0.031
CB892896 similar to GP|10177096|db Notchless protein homolog {Ar... 35 0.068
TC90611 similar to GP|10177223|dbj|BAB10298. contains similarity... 35 0.089
TC91699 similar to GP|21592393|gb|AAM64344.1 unknown {Arabidopsi... 34 0.15
TC84814 homologue to PIR|C86239|C86239 protein T10O24.21 [import... 34 0.15
TC88815 similar to GP|16323464|gb|AAL15226.1 unknown protein {Ar... 34 0.15
AW696217 similar to PIR|T48933|T4 WD repeat domain protein - Ara... 34 0.15
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 33 0.20
BG648991 similar to PIR|C86239|C86 protein T10O24.21 [imported] ... 33 0.20
AW208124 weakly similar to PIR|T49082|T490 hypothetical protein ... 33 0.26
BQ750869 weakly similar to GP|21205303|db alpha-acetolactate syn... 32 0.76
CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~simila... 32 0.76
>TC85306 similar to GP|21593320|gb|AAM65269.1 unknown {Arabidopsis
thaliana}, partial (24%)
Length = 876
Score = 328 bits (841), Expect = 3e-90
Identities = 184/263 (69%), Positives = 204/263 (76%), Gaps = 14/263 (5%)
Frame = +2
Query: 2 MSLISQNARPKIEEK--------EVVDREENSDVDTVKSKKRKRDRKREEHVVDMVEQVR 53
MSLISQNAR K E K EV D +E DVDTV++KKRK D K E+ EQV+
Sbjct: 104 MSLISQNARSKNETKREEVVKGNEVEDGKE-FDVDTVEAKKRKLDGKEEQLAK---EQVK 271
Query: 54 EMRKLESFLFGSLYSPLESGKEVDGEVEP------SDLFFTDRSADSVLSVCDEDGEFSD 107
EM+KLESFLFGSLYSP E GK D EV+ S+LFFTDRSADSVL+V ED +FSD
Sbjct: 272 EMKKLESFLFGSLYSPPEFGKGDDDEVDAAASATASNLFFTDRSADSVLTVYQEDADFSD 451
Query: 108 GSGDGDDGLGRKPAWVDEEEEKFTVNIAKVNRLRKLRKDEDESFISGSEYVARLRAHHVK 167
S D DD L RKP WVDEEEEK TVNIAKVNRLRKLRK+EDE ISGSEYV+RLRA H K
Sbjct: 452 ES-DNDDALKRKPVWVDEEEEKVTVNIAKVNRLRKLRKEEDEDLISGSEYVSRLRAQHAK 628
Query: 168 LNRGTDWAQLDSRLKLDRSDYDGELTDDENEAVVRRGYENVDDILRTNEDLVANSSSKLL 227
LNRGTDWAQLDSR K+D S D ELTDDEN+A V RGYE++DDILRTNEDLV SSSKLL
Sbjct: 629 LNRGTDWAQLDSRSKMDGSS-DDELTDDENKAAVSRGYEDLDDILRTNEDLVVKSSSKLL 805
Query: 228 PGHLEYSRLVDANIQDPSNGPVN 250
PGHLEYS+LVDAN+QDP+NGP+N
Sbjct: 806 PGHLEYSKLVDANVQDPANGPIN 874
>TC78974 similar to GP|22137238|gb|AAM91464.1 AT5g14050/MUA22_5 {Arabidopsis
thaliana}, partial (40%)
Length = 774
Score = 312 bits (799), Expect = 2e-85
Identities = 149/179 (83%), Positives = 167/179 (93%)
Frame = +3
Query: 327 IGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSE 386
IGPL+GREEKS+EVF++S DSQ+IAFVGNEGYILLVSTK+KQLVG+LKM+GT RSLAF+E
Sbjct: 3 IGPLLGREEKSIEVFDVSPDSQLIAFVGNEGYILLVSTKSKQLVGTLKMNGTVRSLAFTE 182
Query: 387 DGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVY 446
DGQKLLSAGGDG +YHWDLRTRTCIHKGVDEGC+N TALCTSP GT FAAGS SG+VN+Y
Sbjct: 183 DGQKLLSAGGDGHIYHWDLRTRTCIHKGVDEGCLNGTALCTSPVGTHFAAGSASGVVNIY 362
Query: 447 NREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSSMKKSSLKLIHIPSYTVFSN 505
N E+FLGGKRKPIKTI+NL T+VDFM+FNHDSQILAICS K SSLKL+HIPSYTVFSN
Sbjct: 363 NSEEFLGGKRKPIKTIDNLNTEVDFMKFNHDSQILAICSRAKNSSLKLVHIPSYTVFSN 539
>TC83040 similar to GP|21593320|gb|AAM65269.1 unknown {Arabidopsis
thaliana}, partial (40%)
Length = 796
Score = 250 bits (638), Expect(2) = 7e-69
Identities = 129/174 (74%), Positives = 143/174 (82%)
Frame = +2
Query: 126 EEEKFTVNIAKVNRLRKLRKDEDESFISGSEYVARLRAHHVKLNRGTDWAQLDSRLKLDR 185
++ K VNIAKVNRLRKLRK+EDE+ ISGSEYV+RLRA HVKLNRGTDWAQLDS K+D
Sbjct: 68 KKRKSPVNIAKVNRLRKLRKEEDENVISGSEYVSRLRAQHVKLNRGTDWAQLDSGSKMDG 247
Query: 186 SDYDGELTDDENEAVVRRGYENVDDILRTNEDLVANSSSKLLPGHLEYSRLVDANIQDPS 245
S D ELTDD N +V RGYE++DDILRTNEDLV SSSKLLPGH+EYS+LVDANIQDPS
Sbjct: 248 SS-DDELTDDGNNVLVSRGYEDLDDILRTNEDLVVKSSSKLLPGHIEYSKLVDANIQDPS 424
Query: 246 NGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFL 299
NGP+NSVQFHRN QLLL AGLD+ LRFFQIDGKRNTKI P K+ FL
Sbjct: 425 NGPINSVQFHRNGQLLLAAGLDRNLRFFQIDGKRNTKISKHLP*RLPHSKSIFL 586
Score = 137 bits (344), Expect = 1e-32
Identities = 68/87 (78%), Positives = 78/87 (89%)
Frame = +3
Query: 282 KIQSIFLEDCPIRKASFLPDGSQVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVF 341
K QSIFLEDCPIRKASFLPDGSQVI+SGRRK FYS DL K ++KIGPL+ REEKS+EVF
Sbjct: 534 KSQSIFLEDCPIRKASFLPDGSQVIISGRRKSFYSLDLFKARVDKIGPLLDREEKSIEVF 713
Query: 342 ELSSDSQMIAFVGNEGYILLVSTKTKQ 368
++S DS+MIAFVGNE +ILLVSTK+KQ
Sbjct: 714 DVSPDSKMIAFVGNERHILLVSTKSKQ 794
Score = 29.3 bits (64), Expect(2) = 7e-69
Identities = 11/14 (78%), Positives = 12/14 (85%)
Frame = +1
Query: 118 RKPAWVDEEEEKFT 131
+KP WVDEEEEK T
Sbjct: 43 KKPVWVDEEEEKVT 84
>BM813821 PIR|T09297|T09 tonoplast intrinsic protein homolog MSMCP1 -
alfalfa, partial (21%)
Length = 641
Score = 48.1 bits (113), Expect = 8e-06
Identities = 26/43 (60%), Positives = 32/43 (73%)
Frame = +1
Query: 32 SKKRKRDRKREEHVVDMVEQVREMRKLESFLFGSLYSPLESGK 74
SKK+K++ E + EQV+EM+KLESFLFGSLYSP E GK
Sbjct: 397 SKKKKKNSN*FEQLAK--EQVKEMKKLESFLFGSLYSPPEFGK 519
Score = 39.3 bits (90), Expect = 0.004
Identities = 28/78 (35%), Positives = 42/78 (52%), Gaps = 6/78 (7%)
Frame = +2
Query: 30 VKSKKRKRDRKREEHVVDMVEQVREMRKLESFLFGSLYSPLESGKEVDGEVEP------S 83
V S+K+K+ + + ++++ + L +F G L GK D EV+ S
Sbjct: 389 VLSQKKKKKTRTSSNSWQR-NKLKK*KNLRAFCLGLSIHLLNLGKGDDDEVDAAASATAS 565
Query: 84 DLFFTDRSADSVLSVCDE 101
+LFFTDRSADSVL+V E
Sbjct: 566 NLFFTDRSADSVLTVYQE 619
>TC84321 GP|22138476|gb|AAM93460.1 hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 748
Score = 43.5 bits (101), Expect = 2e-04
Identities = 22/71 (30%), Positives = 35/71 (48%), Gaps = 3/71 (4%)
Frame = +2
Query: 381 SLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCT---SPSGTLFAAG 437
SL+ S DG++L+SAG D + W L R+C + + +C+ S G +G
Sbjct: 491 SLSLSPDGRELISAGHDASLRFWSLEKRSCTQEITSHRIMRGEGVCSVVWSQDGKWVVSG 670
Query: 438 SDSGIVNVYNR 448
G+V V+ R
Sbjct: 671 GGDGVVKVFAR 703
>TC80911 similar to GP|3668118|emb|CAA11819.1 hypothetical protein {Brassica
napus}, partial (22%)
Length = 860
Score = 42.0 bits (97), Expect = 6e-04
Identities = 38/164 (23%), Positives = 68/164 (41%), Gaps = 11/164 (6%)
Frame = +2
Query: 261 LLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGRRKFFYSFDLV 320
+L+ L+ + + K ++ S+ ED P+ S+ PDGS++ + +D +
Sbjct: 320 VLSIALENDVYLWNASNKSTAELVSVDEEDGPVTSVSWCPDGSRLAIGLDSSLVQVWDTI 499
Query: 321 KG---TLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLK-MS 376
T K G G + ++S ++ G G I+ + + + S + +
Sbjct: 500 ANKQLTTLKSGHRAGVSSLAW------NNSHILTTGGMNGKIVNNDVRVRSHINSYRGHT 661
Query: 377 GTTRSLAFSEDGQKLLSAGGDGQVYHWD-------LRTRTCIHK 413
L +S DG+KL S G D V+ WD RT +HK
Sbjct: 662 DEVCGLKWSLDGKKLASGGSDNVVHIWDRSAVSSSSRTTRWLHK 793
>TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein homolog
{Arabidopsis thaliana}, partial (47%)
Length = 873
Score = 39.7 bits (91), Expect = 0.003
Identities = 25/63 (39%), Positives = 33/63 (51%), Gaps = 2/63 (3%)
Frame = +3
Query: 381 SLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCT--SPSGTLFAAGS 438
S+AFS DG++L S GD V WDL T+T ++ + LC SP G +GS
Sbjct: 510 SVAFSPDGRQLASGSGDTTVRFWDLGTQTPMYTCTGH---KNWVLCIAWSPDGKYLVSGS 680
Query: 439 DSG 441
SG
Sbjct: 681 MSG 689
>AA661007 similar to GP|21539573|gb| WD40-repeat protein {Arabidopsis
thaliana}, partial (8%)
Length = 605
Score = 36.2 bits (82), Expect = 0.031
Identities = 27/78 (34%), Positives = 36/78 (45%), Gaps = 1/78 (1%)
Frame = +1
Query: 364 TKTKQLVGSL-KMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINS 422
TK K + +L S+A SEDG LLSAG D V WDL + + + S
Sbjct: 106 TKRKNCIATLDNHRSAVTSIAVSEDGWTLLSAGRDKVVTLWDLHDYSNKKTVITNEAVES 285
Query: 423 TALCTSPSGTLFAAGSDS 440
+C G+ FA+ DS
Sbjct: 286 --VCAIGVGSPFASSLDS 333
>AL367238 similar to GP|10177204|dbj contains similarity to unknown
protein~gb|AAF44992.1~gene_id:MVP7.5 {Arabidopsis
thaliana}, partial (22%)
Length = 240
Score = 36.2 bits (82), Expect = 0.031
Identities = 16/34 (47%), Positives = 20/34 (58%)
Frame = +2
Query: 380 RSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHK 413
R + + D KL S GGD QVY+WD+ T I K
Sbjct: 11 RDVHVTTDNSKLCSCGGDRQVYYWDVATGRVIRK 112
>CB892896 similar to GP|10177096|db Notchless protein homolog {Arabidopsis
thaliana}, partial (21%)
Length = 850
Score = 35.0 bits (79), Expect = 0.068
Identities = 16/29 (55%), Positives = 20/29 (68%)
Frame = +1
Query: 381 SLAFSEDGQKLLSAGGDGQVYHWDLRTRT 409
S+AFS DG++L S GD V WDL T+T
Sbjct: 709 SVAFSPDGRQLASGSGDTTVRFWDLGTQT 795
>TC90611 similar to GP|10177223|dbj|BAB10298. contains similarity to
GTP-binding regulatory protein and WD-repeat
protein~gene_id:MPF21.14, partial (40%)
Length = 855
Score = 34.7 bits (78), Expect = 0.089
Identities = 38/170 (22%), Positives = 70/170 (40%), Gaps = 11/170 (6%)
Frame = +1
Query: 334 EEKSLEVFELSSDSQMIAFV------GNEGYILLVSTKTKQLVGSLKMS-----GTTRSL 382
+++ + V++L++D V G+ +L+ Q+ K + T SL
Sbjct: 322 QDRKIRVWKLTNDQTKYTHVATLPTLGDRASKILIPKNHVQIRRHKKCTWVHHVDTVSSL 501
Query: 383 AFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGI 442
A S+DG+ L S D + W + TC+ + A+ S GT ++ +D I
Sbjct: 502 ALSKDGKFLYSVSWDRTIKIWRTKDFTCLESITNAHDDAINAVVVSYDGTFYSGSADKRI 681
Query: 443 VNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSSMKKSSL 492
N + K+K ++ + + K H+S I A+ S +S L
Sbjct: 682 KVWKNLQGDDHNKKKHKHSLVDTLEK-------HNSGINALVLSSDESIL 810
>TC91699 similar to GP|21592393|gb|AAM64344.1 unknown {Arabidopsis
thaliana}, partial (71%)
Length = 1035
Score = 33.9 bits (76), Expect = 0.15
Identities = 36/138 (26%), Positives = 62/138 (44%), Gaps = 8/138 (5%)
Frame = +3
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSD--SQMIAFVGNEGYILLVSTKT 366
G K F S KG + LVG + + + S+D S +++ N Y+L
Sbjct: 291 GAIKLFDSRSYDKGPFDTF--LVGGDTAEVCDIKFSNDGKSMLLSTTNNNIYVLDAYGGD 464
Query: 367 KQLVGSLKMS-GTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINS--- 422
K+ SL+ S GT+ F+ DG+ ++++ G G ++ W I + + C +S
Sbjct: 465 KRCGFSLEPSHGTSIEATFTPDGKYVVASSGGGTMHAWS------IDRNHEVACWSSHIG 626
Query: 423 --TALCTSPSGTLFAAGS 438
+ L +P +FAA S
Sbjct: 627 VPSCLKWAPRRAMFAAAS 680
>TC84814 homologue to PIR|C86239|C86239 protein T10O24.21 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 671
Score = 33.9 bits (76), Expect = 0.15
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 6/51 (11%)
Frame = +2
Query: 375 MSGTTRSLAFSEDGQKLLSAGGDGQVYHWD------LRTRTCIHKGVDEGC 419
++G + FS DG+ ++S G+G+ + WD RT C H+GV GC
Sbjct: 14 VAGYACQVNFSPDGRFVMSGDGEGKCWFWDWKSCKVFRTLKC-HEGVTIGC 163
>TC88815 similar to GP|16323464|gb|AAL15226.1 unknown protein {Arabidopsis
thaliana}, partial (45%)
Length = 788
Score = 33.9 bits (76), Expect = 0.15
Identities = 42/190 (22%), Positives = 80/190 (42%), Gaps = 13/190 (6%)
Frame = +1
Query: 236 LVDANIQDPSNGP-VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIR 294
L+D ++ P + + +++F +L ++AG D+ L+ + + N K S L + +
Sbjct: 199 LLDESVNQPFHKDNIRAIRFGAKGKLFVSAGDDKTLKIWSPE---NWKCISTVLSEKRVT 369
Query: 295 KASFLPDGSQVILSGRRKFFYSFDLVKGTLEKIG-PLVGREEKSLEVFELSSDSQMIAFV 353
+ DG V + + + DL K + +K PL+ + E S D++ I
Sbjct: 370 AVAISNDGLYVCFADKFGLVWIVDLNKNSHDKKPIPLLSHYCSIITSLEFSPDNRYILSA 549
Query: 354 GNEGYILLVSTKTKQLVGSLKMS----GTTR---SLAFSEDGQK----LLSAGGDGQVYH 402
+ I + + L G+ ++ G T LAF + LLS GD V
Sbjct: 550 DRDFKIRVTNFPKNPLNGAHEIQSFCLGHTEFVSCLAFVPAQENPHSLLLSGSGDSTVRL 729
Query: 403 WDLRTRTCIH 412
WD+ + ++
Sbjct: 730 WDITSGALLY 759
>AW696217 similar to PIR|T48933|T4 WD repeat domain protein - Arabidopsis
thaliana, partial (11%)
Length = 541
Score = 33.9 bits (76), Expect = 0.15
Identities = 26/93 (27%), Positives = 43/93 (45%)
Frame = +2
Query: 353 VGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIH 412
VG E L + TK + M G+ ++ KLL G +Y WDL +RTC+
Sbjct: 119 VGCEDGCLQIYTKCGRRAMPTMMMGSAAIFVDCDESWKLLLVTRKGSLYLWDLFSRTCL- 295
Query: 413 KGVDEGCINSTALCTSPSGTLFAAGSDSGIVNV 445
+ + ++ A +SPS ++ D+G + V
Sbjct: 296 --LQDSLVSLVA--SSPS----SSAKDTGTIKV 370
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 33.5 bits (75), Expect = 0.20
Identities = 33/106 (31%), Positives = 46/106 (43%), Gaps = 7/106 (6%)
Frame = +1
Query: 373 LKMSGTTRS---LAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINS-TALCTS 428
L G +RS L F DG S G D WDLRT + EG + S + S
Sbjct: 1309 LLQEGHSRSVYGLDFHHDGSLAASCGLDALARVWDLRTGRSVL--ALEGHVKSILGISFS 1482
Query: 429 PSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTI---ENLITKVDF 471
P+G A G + +++ K+K + TI NLI++V F
Sbjct: 1483 PNGYHLATGGEDNTCRIWDLR-----KKKSLYTIPAHSNLISQVKF 1605
>BG648991 similar to PIR|C86239|C86 protein T10O24.21 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 702
Score = 33.5 bits (75), Expect = 0.20
Identities = 14/32 (43%), Positives = 18/32 (55%)
Frame = +3
Query: 376 SGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRT 407
S R + F+ DG K LSAG D + +WD T
Sbjct: 459 SKAVRDICFTNDGTKFLSAGYDKNIKYWDTET 554
>AW208124 weakly similar to PIR|T49082|T490 hypothetical protein F4F15.140 -
Arabidopsis thaliana, partial (19%)
Length = 451
Score = 33.1 bits (74), Expect = 0.26
Identities = 30/128 (23%), Positives = 52/128 (40%), Gaps = 27/128 (21%)
Frame = +2
Query: 308 SGRRKFFYSFD--LVKGTLEKI---GPLVGREEKSLEVFELSS----------------- 345
+G+R F S + +KG+ + +VG ++ S+ VF++ S
Sbjct: 32 NGKRSVFPSLEGKFIKGSCMRYFDPEAMVGCDDGSVRVFDMYSRRCSQIIRMHSAPITCL 211
Query: 346 ---DSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTT--RSLAFSEDGQKLLSAGGDGQV 400
+ Q+I G I + + Q V +L+ S ++L Q L + G
Sbjct: 212 CLSEDQLILSGSTSGNITIADPSSVQKVATLRSSDFRGIKTLCLKPSSQLLFAGSAVGYT 391
Query: 401 YHWDLRTR 408
Y WD+RTR
Sbjct: 392 YCWDMRTR 415
>BQ750869 weakly similar to GP|21205303|db alpha-acetolactate synthase
{Staphylococcus aureus subsp. aureus MW2}, partial (18%)
Length = 732
Score = 31.6 bits (70), Expect = 0.76
Identities = 17/49 (34%), Positives = 26/49 (52%)
Frame = -1
Query: 354 GNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYH 402
G+EG +LLV+ K +G + + D + +L AGGD +VYH
Sbjct: 222 GDEGGVLLVAAHDKLNLGPVDQGVEDGVDLGARDAKDVLDAGGDERVYH 76
>CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~similar to unknown
protein~sp|O15736 {Arabidopsis thaliana}, partial (59%)
Length = 916
Score = 31.6 bits (70), Expect = 0.76
Identities = 18/78 (23%), Positives = 32/78 (40%)
Frame = +3
Query: 330 LVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQ 389
L G +K V S+++ +G I + ++ +L FS DGQ
Sbjct: 480 LTGHTDKVCAVDISKVSSRLVVSAAYDGTIKVWDLMKGYCTNTIMFPSICNALCFSTDGQ 659
Query: 390 KLLSAGGDGQVYHWDLRT 407
+ S DG + WD+++
Sbjct: 660 TIFSGHVDGNLRLWDIQS 713
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.134 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,888,570
Number of Sequences: 36976
Number of extensions: 162334
Number of successful extensions: 815
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 784
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 806
length of query: 509
length of database: 9,014,727
effective HSP length: 100
effective length of query: 409
effective length of database: 5,317,127
effective search space: 2174704943
effective search space used: 2174704943
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0055.8


